Difference between revisions of "HAEM5:Myeloid proliferations associated with Down syndrome"
| [checked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Myeloid proliferations associated with Down syndrome}} | {{DISPLAYTITLE:Myeloid proliferations associated with Down syndrome}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
{{Under Construction}} | {{Under Construction}} | ||
| − | <blockquote class='blockedit'>{{Box-round|title= | + | <blockquote class='blockedit'>{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome]]. |
Other relevent pages include: [[HAEM4:Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome]], [[HAEM4:Myeloid Leukemia Associated with Down Syndrome]] | Other relevent pages include: [[HAEM4:Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome]], [[HAEM4:Myeloid Leukemia Associated with Down Syndrome]] | ||
| Line 10: | Line 10: | ||
}}</blockquote> | }}</blockquote> | ||
| − | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples). Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> |
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| Line 43: | Line 43: | ||
==Clinical Features== | ==Clinical Features== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |EXAMPLE Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | EXAMPLE B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | EXAMPLE Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | EXAMPLE Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |EXAMPLE Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | EXAMPLE Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 82: | Line 82: | ||
==Immunophenotype== | ==Immunophenotype== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 88: | Line 88: | ||
!Finding!!Marker | !Finding!!Marker | ||
|- | |- | ||
| − | |Positive (universal)||EXAMPLE CD1 | + | |Positive (universal)||<span class="blue-text">EXAMPLE:</span> CD1 |
|- | |- | ||
| − | |Positive (subset)||EXAMPLE CD2 | + | |Positive (subset)||<span class="blue-text">EXAMPLE:</span> CD2 |
|- | |- | ||
| − | |Negative (universal)||EXAMPLE CD3 | + | |Negative (universal)||<span class="blue-text">EXAMPLE:</span> CD3 |
|- | |- | ||
| − | |Negative (subset)||EXAMPLE CD4 | + | |Negative (subset)||<span class="blue-text">EXAMPLE:</span> CD4 |
|} | |} | ||
| Line 117: | Line 117: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 150: | Line 150: | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain / Loss / LOH== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 160: | Line 160: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |EXAMPLE Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 173: | Line 173: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |EXAMPLE Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 190: | Line 190: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 202: | Line 202: | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| − | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis'')</span> | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 212: | Line 212: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 218: | Line 218: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 230: | Line 230: | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV / INDEL)== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 240: | Line 240: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE: TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | EXAMPLE: | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | EXAMPLE: BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |EXAMPLE: TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |EXAMPLE: 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE: 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |EXAMPLE: IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |EXAMPLE: EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 282: | Line 282: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table.'')</span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |EXAMPLE: BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |EXAMPLE: MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |EXAMPLE: Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |EXAMPLE: CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |EXAMPLE: Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |EXAMPLE: Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |EXAMPLE: KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |EXAMPLE: Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |EXAMPLE: Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
Revision as of 17:05, 6 September 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome.Other relevent pages include: HAEM4:Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome, HAEM4:Myeloid Leukemia Associated with Down Syndrome
Note: author needs to merge content from Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome AND Myeloid Leukemia Associated with Down Syndrome
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Linda D Cooley, MD, MBA, Children's Mercy Hospital, Kansas City, MO
Cancer Category / Type
HAEM4:Myeloid Proliferations Associated with Down Syndrome
Cancer Sub-Classification / Subtype
Transient abnormal myelopoiesis (TAM) associated with Down syndrome
Definition / Description of Disease
This is a distinct entity in the World Health Organization (WHO) classification system within the section of HAEM4:Myeloid Proliferations Associated with Down Syndrome[1].
Transient abnormal myelopoiesis (TAM) associated with Down syndrome (DS) is a unique disorder of newborns with DS that presents with clinical and morphological findings indistinguishable from those of acute myeloid leukemia. The blasts have morphological and immunological features of megakaryocytic lineage[1][2][3][4][5][6].
Synonyms / Terminology
Tansient myeloproliferative disorder (TMD)
Epidemiology / Prevalence
TAM is diagnosed in ~10% of newborns with DS, but the true incidence is higher because not all newborns are symptomatic. TAM uncommonly occurs in phenotypically normal neonates with trisomy 21 mosaicism, and is extremely rare in neonates without chromosome 21 abnormalities[1][2][3][4][5][6].
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.Transient abnormal myelopoiesis occurs exclusively in patients with Down syndrome (constitutional trisomy 21), and affects approximately 4% to 10% of neonates. TAM manifests in the neonatal period, and is characterized by circulating megakaryoblasts with varied degrees of multisystem organ involvement. The median age of presentation is 3 to 7 days, though patients may be diagnosed at up to 2 months of life. The most common clinical manifestations include hepatomegaly (60%), splenomegaly (35%–40%), jaundice (15%), pericardial effusion (15%), pleural effusion (10%–15%), ascites (10%), respiratory distress (10%), and bleeding diathesis (10%). Less common features include hepatic fibrosis, hydrops fetalis, and renal failure. Characteristic hematologic findings include leukocytosis (WBC > 100K/μL in 20%–30%), thrombocytopenia (40%), and increased numbers of circulating blasts. Approximately 10% to 25% of patients are asymptomatic; thus, the diagnosis may be established as an incidental finding during laboratory assessment for some other cause. Occasionally, the finding of TAM may even be the first indication that a patient has trisomy 21.
The natural history of TAM is quite variable. Most neonates (80% with documented TAM) undergo spontaneous remission within 3 to 6 months of age. At the other end of the disease spectrum, neonatal (or even fetal) demise occurs in approximately 10% of patients secondary to diffuse organ infiltration by megakaryoblasts, associated parenchymal fibrosis, and subsequent multisystem failure. Independent risk factors that portend early mortality include WBC counts above 100 K/μL, preterm delivery (<37 weeks), effusions (pleural, pericardial, ascites, or hydrops), coagulopathy, bleeding diathesis, platelet count greater than 100K/μL, low birth weight, and failure to clear peripheral blasts. When these high-risk features are present, chemotherapeutic intervention is warranted.
Approximately 20% of patients with TAM develop AMKL within the first 4 years of life and this may be preceded by a myelodysplastic-like syndrome. The World Health Organization classification category “myeloid leukemia associated with DS” (DS-AMKL) encompasses both myelodysplastic and leukemic manifestations regardless of blast percentage. The median age of onset is 2 years, younger than that seen in non–DS-AMKL. Patients typically manifest with low WBC count, cytopenia, organomegaly, progressive marrow fibrosis, and clonal cytogenetic abnormalities, e.g., trisomy 8. Blasts in DS-AMKL are morphologically and immunophenotypically similar to those seen in TAM. Patients with DS-AMKL have a favorable prognosis with 80% 3-year overall survival. This response rate is, in part, attributed to enhanced chemosensitivity of megakaryoblasts to cytarabine. The cytidine deaminase gene functions in cytarabine catabolism, and its transcription is diminished in DS, which may lead to diminished intracellular drug metabolism and consequent increased drug efficacy[1][2][3][4][5].
Sites of Involvement
Blood and bone marrow are the principle sites of involvement. Extramedullary involvement, mainly of the spleen and liver, is almost always present as well[1].
Morphologic Features
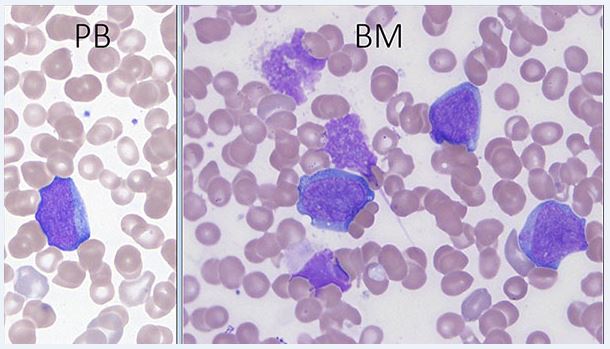
The morphological and immunophenotypic features of TAM are similar to those seen in most cases of acute myeloid leukemia associated with DS[1]. Peripheral blood and bone marrow blasts often have basophilic cytoplasm and coarse basophilic granules and cytoplasmic blebbing suggestive of megakaryoblasts. Some patients have peripheral blood basophilia; erythroid and megakaryocytic dysplasia is often present in the bone marrow. No critical threshold has been established for blast percentage in the diagnosis of TAM. Circulating blasts can often be seen in both neonates without DS, especially those who are ill or premature, and neonates with DS, emphasizing the importance of manual peripheral smear review as well as ancillary laboratory studies in the evaluation [5]. Bone marrow examination may not be indicated as marrow findings are either similar to, or less pronounced than, those in blood[5].
Immunophenotype
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Finding | Marker |
|---|---|
| Positive (universal) | EXAMPLE: CD1 |
| Positive (subset) | EXAMPLE: CD2 |
| Negative (universal) | EXAMPLE: CD3 |
| Negative (subset) | EXAMPLE: CD4 |
editv4:ImmunophenotypeThe content below was from the old template. Please incorporate above.Phenotypically, blasts commonly express stem cell (variable CD34, CD117), myeloid (CD13, CD33), nonlineage (CD4, CD7, CD56), and megakaryoblastic/megakaryocytic (CD61, CD41, CD42) antigens[5].
The blasts in TAM have a characteristic megakaryoblastic immunophenotype. In most cases, the leukemic blasts are positive for CD34, KIT (CD117), CD13, CD33, HLA-DR, CD4 (dim), CD41, CD42, CD110 (TPOR), IL3R, CD36, CD61, and CD71, often with expression of CD7 and CD56. The blasts are negative for MPO, CD15, CD14, CD11a, and glycophorin A. IHC with CD41, CD42b, and CD 61 may be particularly useful for identifying blasts of megakaryocytic lineage in BM biopsies[1].
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.not applicable
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
Transient abnormal myelopoiesis is a preleukemic disorder that occurs only in neonates with constitutional trisomy 21. Transient abnormal myelopoiesis typically presents in the first week of life with leukocytosis, thrombocytopenia, hepatomegaly, and circulating megakaryoblasts, the latter of which contain an acquired GATA1 mutation. Although TAM can be fatal in 10% of patients, it most often resolves spontaneously, but is believed to persist in a “quiescent” state. By 5 years of age, 20% of patients progress to AMKL following an intervening remission and/or a preceding myelodysplastic-like syndrome. Down syndrome–AMKL has a favorable prognosis with enhanced chemotherapeutic responsiveness to cytarabine.
The disease evolution of TAM and DS-AMKL is currently conceived as a sequential multistep process of leukemogenesis. Trisomy 21 represents the critical “initiating” event. GATA1 mutation reflects a “secondary hit” to fetal liver hematopoiesis, particularly megakaryocytic-erythroid progenitors, that leads to TAM and confers some selective advantage.
Given the risk of progression to DS-AMKL, some advocate that all neonates with DS undergo routine screening for TAM with manual peripheral blood smear review and GATA1 mutation analysis. With detection of a GATA1 mutation, clinical assessment and routine laboratory screening is then suggested periodically throughout early childhood[1][5].
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.not applicable
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.Constitutional trisomy 21 or mosaic constitutional trisomy 21
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.
Gene Mutation Oncogene/Tumor Suppressor/Other Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) Prevalence (COSMIC/TCGA/Other) GATA1 Most mutations insert a premature termination codon either by introducing a stop codon or frameshift. Mutations affecting the splice site at GATA1 exon 2 exon/intron boundary are next most frequent. zinc finger DNA-binding transcription factor that plays a critical role in the normal development of hematopoietic cell lineages N-terminally truncating somatic mutation 100% Other Mutations
Not applicable
Epigenomic Alterations
not applicable
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.In addition to trisomy 21, acquired GATA1 mutations are present in blast cells of TAM [1].
Somatic GATA1 mutations are pivotal in the development of TAM and have proven to be a marker of clonal identity in its evolution to DS-AMKL. The GATA1 gene is located on the X chromosome and encodes a zinc finger transcription factor that is essential for normal erythropoiesis and megakaryopoiesis. Its de novo protein product contributes to cytoplasmic maturation in megakaryocytes and organelle development in platelets, but functions as a negative regulator of megakaryocyte proliferation. Various acquired mutations in exon 2, or less commonly exon 3, ultimately yield a mutant N-terminally truncated GATA1 protein (designated GATA1s) that has been detected exclusively in patients with DS. GATA1 mutations (both single and multiple clones) have been detected in Guthrie card blood spots of patients with DS.
In normal human development, the liver is the primary site of fetal (in utero) hematopoiesis. With birth, the hepatic microenvironment changes such that liver hematopoiesis is down-regulated while bone marrow simultaneously assumes this primary functionality. Given the clinical and laboratory manifestations of TAM, which include leukocytosis and circulating megakaryoblasts, often with hepatomegaly, TAM reflects perturbation of this normal developmental process. Transient abnormal myelopoiesis may arise in utero within the fetal liver with “spontaneous resolution” reflecting the natural process of hepatic hematopoietic down-regulation.
TAM is the result of a multistep process in which trisomy 21 is the “initiating” event in disease pathogenesis. Trisomy 21 creates an environment, in utero, in which hematopoietic progenitor cells within fetal liver are primed for acquisition of either single or multiple somatic GATA1 mutations that reflect a “secondary hit,” thereby promoting hematopoietic dysregulation and emergence of TAM. With birth, hematopoiesis naturally transitions from fetal liver to bone marrow and the GATA1 megakaryoblastic clone becomes quiescent. However, this clone persists over time and undergoes other somatic mutations and epigenetic events ultimately lead to the impaired megakaryocytic differentiation and uncontrolled proliferation characteristic of DS-AMKL[2][3][4][5].
Genetic Diagnostic Testing Methods
When TAM is suspected clinically, cytogenetic karyotypic analysis should be performed to establish constitutional trisomy 21, while GATA1 mutation analysis is also recommended to document clonality of the blast population. The presence of an acquired mutation(s) in exon 2 or exon 3 of the GATA1 gene on chromosome X establishes a diagnosis of TAM and serves as a potential marker for future disease monitoring in the development of AMKL. If a GATA1 mutation is detected in a neonate without clinical features of DS, cytogenetic analysis should still be performed to exclude DS mosaicism[5].
Familial Forms
not applicable
Additional Information
Also see HAEM4:Myeloid Proliferations Associated with Down Syndrome
Links
http://www.archivesofpathology.org/doi/pdf/10.5858/arpa.2014-0304-CC?code=coap-site
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2265448/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5031718/pdf/11899_2016_Article_338.pdf
https://link.springer.com/content/pdf/10.1007%2Fs12185-016-1959-5.pdf
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4551356/
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Arber DA, et al., (2017). Myeloid proliferations associated with Down syndrome, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p169-170.
- ↑ 2.0 2.1 2.2 2.3 Klusmann, Jan-Henning; et al. (2008). "Treatment and prognostic impact of transient leukemia in neonates with Down syndrome". Blood. 111 (6): 2991–2998. doi:10.1182/blood-2007-10-118810. ISSN 0006-4971. PMC 2265448. PMID 18182574.
- ↑ 3.0 3.1 3.2 3.3 Gruber, Tanja A.; et al. (2015). "The biology of pediatric acute megakaryoblastic leukemia". Blood. 126 (8): 943–949. doi:10.1182/blood-2015-05-567859. ISSN 1528-0020. PMC 4551356. PMID 26186939.
- ↑ 4.0 4.1 4.2 4.3 Bhatnagar, Neha; et al. (2016). "Transient Abnormal Myelopoiesis and AML in Down Syndrome: an Update". Current Hematologic Malignancy Reports. 11 (5): 333–341. doi:10.1007/s11899-016-0338-x. ISSN 1558-822X. PMC 5031718. PMID 27510823.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Bombery, Melissa; et al. (2014). "Transient abnormal myelopoiesis in neonates: GATA get the diagnosis". Archives of Pathology & Laboratory Medicine. 138 (10): 1302–1306. doi:10.5858/arpa.2014-0304-CC. ISSN 1543-2165. PMID 25268193.
- ↑ 6.0 6.1 Saida, Satoshi (2016). "Evolution of myeloid leukemia in children with Down syndrome". International Journal of Hematology. 103 (4): 365–372. doi:10.1007/s12185-016-1959-5. ISSN 1865-3774. PMID 26910243.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Myeloid proliferations associated with Down syndrome”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 09/6/2024, https://ccga.io/index.php/HAEM5:Myeloid_proliferations_associated_with_Down_syndrome.