Difference between revisions of "HAEM5:Acute myeloid leukaemia with BCR::ABL1 fusion"
| [checked revision] | [checked revision] |
Bailey.Glen (talk | contribs) (Undo revision 14805 by Bailey.Glen (talk)) Tag: Undo |
Bailey.Glen (talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Acute myeloid leukaemia with BCR::ABL1 fusion}} | {{DISPLAYTITLE:Acute myeloid leukaemia with BCR::ABL1 fusion}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
{{Under Construction}} | {{Under Construction}} | ||
| − | <blockquote class='blockedit'>{{Box-round|title= | + | <blockquote class='blockedit'>{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Acute Myeloid Leukemia (AML) with BCR-ABL1]]. |
}}</blockquote> | }}</blockquote> | ||
| − | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples). Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> |
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| Line 15: | Line 15: | ||
__TOC__ | __TOC__ | ||
| − | == | + | ==WHO Classification of Disease== |
| − | + | {| class="wikitable" | |
| − | + | !Structure | |
| − | + | !Disease | |
| − | + | |- | |
| − | + | |Book | |
| + | |Haematolymphoid Tumours (5th ed.) | ||
| + | |- | ||
| + | |Category | ||
| + | |Myeloid proliferations and neoplasms | ||
| + | |- | ||
| + | |Family | ||
| + | |Acute myeloid leukaemia | ||
| + | |- | ||
| + | |Type | ||
| + | |Acute myeloid leukaemia with defining genetic abnormalities | ||
| + | |- | ||
| + | |Subtype(s) | ||
| + | |Acute myeloid leukaemia with BCR::ABL1 fusion | ||
| + | |} | ||
==Definition / Description of Disease== | ==Definition / Description of Disease== | ||
| Line 63: | Line 77: | ||
==Clinical Features== | ==Clinical Features== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |EXAMPLE Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | EXAMPLE B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | EXAMPLE Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | EXAMPLE Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |EXAMPLE Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | EXAMPLE Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 123: | Line 137: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 161: | Line 175: | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain / Loss / LOH== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 171: | Line 185: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |EXAMPLE Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 184: | Line 198: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |EXAMPLE Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 201: | Line 215: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 224: | Line 238: | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| − | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis'')</span> | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 234: | Line 248: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 240: | Line 254: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 252: | Line 266: | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV / INDEL)== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 262: | Line 276: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE: TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | EXAMPLE: | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | EXAMPLE: BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |EXAMPLE: TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |EXAMPLE: 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE: 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |EXAMPLE: IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |EXAMPLE: EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 314: | Line 328: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table.'')</span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |EXAMPLE: BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |EXAMPLE: MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |EXAMPLE: Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |EXAMPLE: CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |EXAMPLE: Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |EXAMPLE: Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |EXAMPLE: KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |EXAMPLE: Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |EXAMPLE: Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
Latest revision as of 17:18, 6 September 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Acute Myeloid Leukemia (AML) with BCR-ABL1.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Kay Weng Choy MBBS, Monash Medical Centre
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Acute myeloid leukaemia |
| Type | Acute myeloid leukaemia with defining genetic abnormalities |
| Subtype(s) | Acute myeloid leukaemia with BCR::ABL1 fusion |
Definition / Description of Disease
AML with BCR-ABL1 is a rare subtype of AML that is now included as a provisional entity under the heading of “AML with recurrent genetic abnormalities” in the 2016 revised World Health Organization (WHO) classification of myeloid malignancies[1]. It must be distinguished from myeloid blast crisis of chronic myeloid leukemia (CML-MBC) and mixed phenotype acute leukemia (MPAL) with BCR-ABL1, which are separate entities in the WHO classification. The t(9:22)(q34.1;q11.2) results in the formation of the Philadelphia chromosome and the chimeric BCR-ABL1 fusion gene. It is worth noting that the fusion gene is the cytogenetic hallmark of chronic myelogenous leukemia (CML) and is frequently found in high-risk acute lymphoblastic leukemia (ALL).
Table [adapted from [2]] - Differentiating CML-MBC from AML with BCR-ABL1
| CML-MBC | AML with BCR-ABL1 |
|---|---|
| Previous blood disorder, idiopathic leucocytosis | No previous blood disorder |
| Splenomegaly | No splenomegaly |
| Basophilia (≥2%) | No basophilia |
| ~100% marrow cellularity | <100% marrow cellularity (median 80%) |
| Typical AML blast crisis phenotype | Aberrant CD19, CD7 and TdT more common |
| Ph+ in ~100% of cells | Ph+ in <100% of cells |
| P210 transcript in >99% of cases | p190 and p210 detected in nearly equal distribution |
| Loss of IKZF1 and CDKN2A and cryptic deletions within IGH and TCR genes |
Most cases which carry the BCR-ABL1 fusion have been described as de novo AML; there are only a few cases described where acquisition of the Philadelphia chromosome upon disease relapse or transformation of antecedent myelodysplastic syndrome into AML occurred[3]. Based on one model of leukemogenesis in which two driver mutations are required (a class-II mutation, conferring inhibition of differentiation and apoptosis, and a class-I mutation conferring a proliferative advantage), BCR-ABL1 is likely to represent a class-I mutation[3].
Synonyms / Terminology
Not applicable
Epidemiology / Prevalence
It is reported that up to 3% of all AML carry BCR-ABL1 fusions. The overall incidence is mainly estimated from larger case series without any standardized definition[3].
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.Little is known about the characteristics clinical features of BCR-ABL1 positive AML. In contrast to CML, BCR-ABL1 positive AML has no antecedent hematologic disease/abnormality and no splenomegaly. Unexplained leucocytosis and/or splenomegaly point towards the diagnosis of CML myeloid blast crisis (CML-MBC) than AML[3]. Soupir et al. reported 16 cases of BCR-ABL1 positive AML, noting there was some overlap phenotypically and morphologically with CML myeloid blast crisis, but BCR-ABL1 positive AML cases presented less often with splenomegaly, lacked basophilia and had lower bone marrow cellularity[4].
Sites of Involvement
Bone marrow
Morphologic Features
BCR-ABL1 positive AML features include myeloid blasts >20% found in the bone marrow, below a blast count of 20% the WHO classification currently describes diagnosis of BCR-ABL1 AML as controversial. It is helpful to note several features that may help in the differential diagnosis of BCR-ABL1 positive AML: basophilia (>2% in white blood cells) strongly diminishes the probability of de novo AML (most cases of BCR-ABL1 positive AML are de novo AML); prior signs of myelodysplastic syndrome (MDS) in peripheral blood or bone marrow may support the diagnosis of secondary AML[3].
Immunophenotype
Co-expression of lymphoid markers have been reported in BCR-ABL1 positive cases. Atfy et al., reported an aberrant lymphoid co-expression in seven of nine BCR-ABL1 positive AML cases[5]. Soupir et al. reported that there was no significant difference in the blast immunophenotype by flow cytometry between the BCR-ABL positive AML and CML-MBC cases. In nearly all cases in both groups, the blasts expressed CD13, CD33, and CD34[4]. However, compared to CML-MBC, aberrant CD19, CD7 and TdT have been found to be more common in BCR-ABL1 positive AML[2].
| Finding | Marker |
|---|---|
| Positive (universal) | Myeloid antigens (CD13 and CD33), CD34 |
| Positive (subset) | CD19, CD7, TdT |
| Negative (universal) | |
| Negative (subset) |
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.The t(9:22)(q34.1;q11.2) results in the formation of the Ph chromosome and the chimeric BCR-ABL1 fusion gene. In AML, the most common BCR-ABL1 transcripts p190 and p210 have been detected in nearly equal distribution[3]. Since p190 is very rare in CML (p210 transcripts in >99% of cases), the presentation with a p190 transcript is in favour of the diagnosis of AML rather than CML[2].
Chromosomal Rearrangement Genes in Fusion (5’ or 3’ Segments) Pathogenic Derivative Prevalence t(9;22)(q34;q11.2) 3'ABL1 / 5'BCR der(22) <3% of AML
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
BCR-ABL1 positive AML is an emerging entity. The proliferation of BCR-ABL1 positive blasts present a diagnostic dilemma. While it may be difficult, it is essential to distinguish between BCR-ABL1 positive AML and CML-MBC, in order to choose the most appropriate therapy (e.g., intensive induction chemotherapy versus tyrosine kinase inhibitor (TKI) followed by an early allogeneic stem cell transplant). After the exclusion of acute leukemia of ambiguous lineage (a separate entity according to WHO) by flow cytometry, it is helpful to note any past history of antecedent hematological disease. Absence of basophilia and absence of splenomegaly favour the diagnosis of BCR-ABL1 positive AML (over CML-MBC). The detection of p190 transcript and the occurrence of any BCR-ABL1 transcript in less than 100% of metaphases supports the diagnosis of AML rather than CML. Persistent CCyR (Complete Cytogenetic Response) after conventional chemotherapy is unusual for CML-MBC and supports the diagnosis of BCR-ABL1 positive AML[3].
The overall prognosis of BCR-ABL1 positive AML is generally unfavourable. The National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology for AML categorise this entity into the poor-risk group, comparable with complex aberrant karyotype AML[6]. It appears that the prognosis of BCR-ABL1 positive AML depends more on the genetic background (concurrent aberrations) than on BCR-ABL1 itself. Unlike in CML, BCR-ABL1 does not appear to be the key driver in AML though may provide a proliferative advantage to a particular BCR-ABL1 positive subclone There is currently no standardized treatment approach for BCR-ABL1 positive AML. Therapy with TKI alone does not produce sustained responses in BCR-ABL1 positive AML. This may be due to a very rapid clonal evolution, resulting in resistance in a much higher proportion of patients and in a significantly shorter time than in CML[3].
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.AML with BCR-ABL1 carries unique genome imbalances. Nacheva et al., used array comparative genomic hybridisation (CGH) to perform a comparative study between several BCR-ABL1 positive entities. BCR-ABL1 positive AML displays characteristic of lymphoid disease (found in BCR-ABL1 positive ALL and CML): deletions of IKZF1 and/or CDKN2A/B genes were recurrent findings in BCR-ABL1 positive AML as well as cryptic deletions within the immunoglobulin IGH and T cell receptor gene (TRG alpha) complexes[7]. Importantly, these aberrations were found to be absent in CML-MBC and hence they are potentially a helpful diagnostic tool for difficult cases.
Most cases will have monosomy 7, trisomy 8 or complex karyotypes in addition to the t(9;22)(q34.1;q11.2)[1].
Chromosome Number Gain/Loss/Amp/LOH Region 7 Loss chr7 8 Gain chr8
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.In AML, BCR-ABL1 has been described together with different class II aberrations such as CBFB-MYH11, RUNX1- RUNX1T1 and PML-RARA[3]. In AML, BCR-ABL1 seems to cooperate with several AML-specific aberrations such as inv(16), t(8;21) and myelodysplasia-related cytogenetic aberrations[3][8]. (For diagnostic purpose, note that inv(16) is not restricted to AML and can also be found in CML-MBC).
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.Coinciding molecular events such as NPM1 mutations have been reported[2].
Gene Mutation Oncogene/Tumor Suppressor/Other Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) Prevalence (COSMIC/TCGA/Other) NPM1 Other Mutations
Type Gene/Region/Other Concomitant Mutations Secondary Mutations Mutually Exclusive
Epigenomic Alterations
Not applicable
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.The BCR gene product has serine/threonine kinase activity and is a GTPase-activating protein for p21rac[9]. The ABL1 gene is a proto-oncogene that encodes a protein tyrosine kinase involved in a variety of cellular processes, including cell division, adhesion, differentiation, and response to stress. The activity of this protein is negatively regulated by its SH3 domain, whereby deletion of the region encoding this domain results in an oncogene[10]. The t(9,22)(q34;q11) leads to the formation of a Philadelphia chromosome and generates an active chimeric BCR-ABL1 tyrosine kinase. The fusion gene is created by juxtaposing the ABL1 gene on chromosome 9 (region q34) to a part of BCR (breakpoint cluster region) gene on chromosome 22 (region q11). This is a reciprocal translocation, creating an elongated chromosome 9 (der 9), and a truncated chromosome 22 (the Philadelphia chromosome, 22q-), the oncogenic BCR-ABL1 being found on the shorter derivative 22 chromosome[11][12]. This gene encodes for a BCR-ABL1 fusion protein, a tyrosine kinase. Tyrosine kinase activities are typically regulated in an auto-inhibitory manner, but the BCR-ABL1 fusion gene codes for a protein that is continuously activated, causing unregulated cell division. This is a result of the replacement of the myristoylated cap region which causes a conformational change rendering the kinase domain inactive, with a truncated portion of the BCR protein[13]. The enzyme is responsible for the uncontrolled growth of leukemic cells which survive better than normal blood cells. As a result of BCR/ABL1 variable splicing (fusion RNA and hybrid proteins), two transcripts p190 and p210 are found for BCR-ABL1 positive AML.
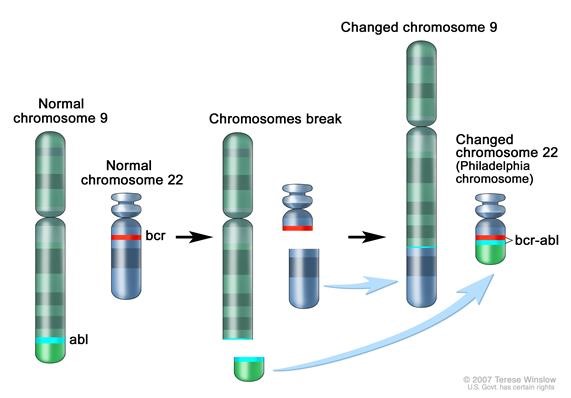 Figure 1. Philadelphia chromosome. A piece of chromosome 9 and a piece of chrosomome 22 break off and trade places. The BCR-ABL1 gene is formed on chromosome 22 where the piece of chromosome 9 attaches. The changed chromosome 22 is called Philadelphia chromosome. Image from National Cancer Institute website https://www.cancer.gov/publications/dictionaries/cancer-terms/def/bcr-abl-fusion-gene
Figure 1. Philadelphia chromosome. A piece of chromosome 9 and a piece of chrosomome 22 break off and trade places. The BCR-ABL1 gene is formed on chromosome 22 where the piece of chromosome 9 attaches. The changed chromosome 22 is called Philadelphia chromosome. Image from National Cancer Institute website https://www.cancer.gov/publications/dictionaries/cancer-terms/def/bcr-abl-fusion-gene
Genetic Diagnostic Testing Methods
Bone marrow with myeloid blasts >20% combined with detection of t(9,22) by karytoype analysis or BCR-ABL1 using FISH or reverse transcriptase-quantitative PCR (RT-qPCR)[3]. A graphic of the clinical path for the differential diagnosis of BCR-ABL1 positive acute myeloid leukemia and chronic myeloid leukemia-myeloid blast crisis (CML-MBC) is presented[3].
Familial Forms
Not applicable
Additional Information
Not applicable
Links
Vogel A, et al. Acute Myeloid Leukemia with BCR-ABL1. SH2017-0299. Presentation at Society for Hematopathology / European Association for Haematopathology (SH/EAHP) 2017 Workshop. https://www.sh-eahp.org/images/2017_Workshop/3_3SH-EAHP%202017_AML%20with%20BCR-ABL1_%20AV%20FINAL.pdf (accessed 29th June 2018)
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. Revised 4th Edition. IARC Press: Lyon, France, p140.
- ↑ 2.0 2.1 2.2 2.3 Arber DA, et al., (2017). Acute Myeloid Leukemia, in Hematopathology, 2nd Edition. Jaffe E, Arer DA, Campo E, Harris NL, and Quintanilla-Fend L, Editors. Elsevier:Philadelphia, PA, p817-846.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 Neuendorff, Nina Rosa; et al. (2016). "BCR-ABL-positive acute myeloid leukemia: a new entity? Analysis of clinical and molecular features". Annals of Hematology. 95 (8): 1211–1221. doi:10.1007/s00277-016-2721-z. ISSN 1432-0584. PMID 27297971.
- ↑ 4.0 4.1 Soupir, Chad P.; et al. (2007). "Philadelphia chromosome-positive acute myeloid leukemia: a rare aggressive leukemia with clinicopathologic features distinct from chronic myeloid leukemia in myeloid blast crisis". American Journal of Clinical Pathology. 127 (4): 642–650. doi:10.1309/B4NVER1AJJ84CTUU. ISSN 0002-9173. PMID 17369142.
- ↑ Atfy, Maha; et al. (2011). "Incidence of Philadelphia-chromosome in acute myelogenous leukemia and biphenotypic acute leukemia patients: And its role in their outcome". Leukemia Research. 35 (10): 1339–1344. doi:10.1016/j.leukres.2011.04.011. ISSN 1873-5835. PMID 21612824.
- ↑ O’Donnell MR, Tallman MS, (2016). NCCN Clinical Practise Guidelines in Oncology: AML. Version 1. Available at: NCCN.org.
- ↑ Nacheva, Ellie P.; et al. (2013). "Does BCR/ABL1 positive acute myeloid leukaemia exist?". British Journal of Haematology. 161 (4): 541–550. doi:10.1111/bjh.12301. ISSN 1365-2141. PMID 23521501.
- ↑ Bacher, Ulrike; et al. (2011). "Subclones with the t(9;22)/BCR-ABL1 rearrangement occur in AML and seem to cooperate with distinct genetic alterations". British Journal of Haematology. 152 (6): 713–720. doi:10.1111/j.1365-2141.2010.08472.x. ISSN 1365-2141. PMID 21275954.
- ↑ Maru, Y.; et al. (1991). "The BCR gene encodes a novel serine/threonine kinase activity within a single exon". Cell. 67 (3): 459–468. doi:10.1016/0092-8674(91)90521-y. ISSN 0092-8674. PMID 1657398.
- ↑ Wang, Jean Y. J. (2014). "The capable ABL: what is its biological function?". Molecular and Cellular Biology. 34 (7): 1188–1197. doi:10.1128/MCB.01454-13. ISSN 1098-5549. PMC 3993570. PMID 24421390.
- ↑ Kurzrock, Razelle; et al. (2003). "Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics". Annals of Internal Medicine. 138 (10): 819–830. doi:10.7326/0003-4819-138-10-200305200-00010. ISSN 1539-3704. PMID 12755554.
- ↑ Melo, J. V. (1996). "The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype". Blood. 88 (7): 2375–2384. ISSN 0006-4971. PMID 8839828.
- ↑ Nagar, Bhushan; et al. (2003). "Structural basis for the autoinhibition of c-Abl tyrosine kinase". Cell. 112 (6): 859–871. doi:10.1016/s0092-8674(03)00194-6. ISSN 0092-8674. PMID 12654251.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Acute myeloid leukaemia with BCR::ABL1 fusion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 09/6/2024, https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_BCR::ABL1_fusion.