Difference between revisions of "HAEM5:Myelodysplastic/myeloproliferative neoplasm with neutrophilia"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Myelodysplastic/myeloproliferative neoplasm with neutrophilia}} | {{DISPLAYTITLE:Myelodysplastic/myeloproliferative neoplasm with neutrophilia}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
{{Under Construction}} | {{Under Construction}} | ||
| − | <blockquote class='blockedit'>{{Box-round|title= | + | <blockquote class='blockedit'>{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Atypical Chronic Myeloid Leukemia (aCML), BCR-ABL1 Negative]]. |
}}</blockquote> | }}</blockquote> | ||
| + | |||
| + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | ||
| + | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| Line 12: | Line 15: | ||
__TOC__ | __TOC__ | ||
| − | == | + | ==WHO Classification of Disease== |
| − | Myeloid | + | {| class="wikitable" |
| − | + | !Structure | |
| − | + | !Disease | |
| − | + | |- | |
| − | + | |Book | |
| + | |Haematolymphoid Tumours (5th ed.) | ||
| + | |- | ||
| + | |Category | ||
| + | |Myeloid proliferations and neoplasms | ||
| + | |- | ||
| + | |Family | ||
| + | |Myelodysplastic/myeloproliferative neoplasms | ||
| + | |- | ||
| + | |Type | ||
| + | |N/A | ||
| + | |- | ||
| + | |Subtype(s) | ||
| + | |Myelodysplastic/myeloproliferative neoplasm with neutrophilia | ||
| + | |} | ||
==Definition / Description of Disease== | ==Definition / Description of Disease== | ||
| Line 24: | Line 41: | ||
Atypical chronic myeloid leukemia, ''BCR-ABL1''-negative (aCML) presents with myelodysplastic as well as myeloproliferative features at the time of disease onset<ref name=":5" /><ref>Czader, M and Orazi, A. Chapter 45. Myelodysplastic/Myeloproliferative Neoplasms and Related Diseases. In Orazi A, Foucar K, Knowles DM. eds. Knowles Neoplastic Hematopathology. Riverwoods, IL: Wolters Kluwer Health; 2013:1148-1150.</ref>. The characteristic finding is blood leukocytosis with increase of dysplastic neutrophils and immature granulocytes. Bone marrow shows multilineage dysplasia. | Atypical chronic myeloid leukemia, ''BCR-ABL1''-negative (aCML) presents with myelodysplastic as well as myeloproliferative features at the time of disease onset<ref name=":5" /><ref>Czader, M and Orazi, A. Chapter 45. Myelodysplastic/Myeloproliferative Neoplasms and Related Diseases. In Orazi A, Foucar K, Knowles DM. eds. Knowles Neoplastic Hematopathology. Riverwoods, IL: Wolters Kluwer Health; 2013:1148-1150.</ref>. The characteristic finding is blood leukocytosis with increase of dysplastic neutrophils and immature granulocytes. Bone marrow shows multilineage dysplasia. | ||
| − | *Before the diagnosis of aCML can be rendered, myeloid neoplasms with well defined genetic abnormalities such as ''[[Chronic | + | *Before the diagnosis of aCML can be rendered, myeloid neoplasms with well defined genetic abnormalities such as ''[[HAEM5:Chronic myeloid leukaemia|BCR-ABL1]]'' fusion; rearrangement of [[HAEM5:Myeloid/lymphoid neoplasm with PDGFRA rearrangement|P''DGFRA'']]'', [[HAEM5:Myeloid/lymphoid neoplasm with PDGFRB rearrangement|PDGFRB]],'' ''[[HAEM5:Myeloid/lymphoid neoplasm with FGFR1 rearrangement|FGFR1]]'', or ''[[HAEM5:Myeloid/lymphoid neoplasm with JAK2 rearrangement|PCM1-JAK2]]'' must be ruled out. |
| − | *Myeloid neoplasm with features of aCML developed after chemo or radiation therapy is classified as [[ | + | *Myeloid neoplasm with features of aCML developed after chemo or radiation therapy is classified as [[HAEM5:Myeloid neoplasm post cytotoxic therapy|therapy-related myeloid neoplasm]]. |
==Synonyms / Terminology== | ==Synonyms / Terminology== | ||
| Line 34: | Line 51: | ||
==Epidemiology / Prevalence== | ==Epidemiology / Prevalence== | ||
| − | The incidence of aCML is low; it is estimated that there are 1-2 aCML cases for every 100 cases of [[Chronic | + | The incidence of aCML is low; it is estimated that there are 1-2 aCML cases for every 100 cases of [[HAEM5:Chronic myeloid leukaemia|''BCR-ABL1''-positive chronic myeloid leukemia]]<ref>{{Cite journal|last=A|first=Orazi|last2=U|first2=Germing|date=2008|title=The Myelodysplastic/Myeloproliferative Neoplasms: Myeloproliferative Diseases With Dysplastic Features|url=https://pubmed.ncbi.nlm.nih.gov/18480833/|language=en|pmid=18480833}}</ref><ref name=":0">{{Cite journal|last=Sa|first=Wang|last2=Rp|first2=Hasserjian|last3=Ps|first3=Fox|last4=Hj|first4=Rogers|last5=Jt|first5=Geyer|last6=D|first6=Chabot-Richards|last7=E|first7=Weinzierl|last8=J|first8=Hatem|last9=J|first9=Jaso|date=2014|title=Atypical Chronic Myeloid Leukemia Is Clinically Distinct From Unclassifiable Myelodysplastic/Myeloproliferative Neoplasms|url=https://pubmed.ncbi.nlm.nih.gov/24627528/|language=en|doi=10.1182/blood-2014-02-553800|pmc=PMC4067498|pmid=24627528}}</ref>. |
The median patient age at diagnosis is in the seventh or eighth decade of life<ref name=":0" /><ref name=":1">{{Cite journal|last=Jm|first=Hernández|last2=Mc|first2=del Cañizo|last3=A|first3=Cuneo|last4=Jl|first4=García|last5=Nc|first5=Gutiérrez|last6=M|first6=González|last7=G|first7=Castoldi|last8=Jf|first8=San Miguel|date=2000|title=Clinical, Hematological and Cytogenetic Characteristics of Atypical Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/10847463/|language=en|pmid=10847463}}</ref><ref name=":2">{{Cite journal|last=P|first=Martiat|last2=Jl|first2=Michaux|last3=J|first3=Rodhain|date=1991|title=Philadelphia-negative (Ph-) Chronic Myeloid Leukemia (CML): Comparison With Ph+ CML and Chronic Myelomonocytic Leukemia. The Groupe Français De Cytogénétique Hématologique|url=https://pubmed.ncbi.nlm.nih.gov/2070054/|language=en|pmid=2070054}}</ref>. Rare cases of aCML has also been reported in teenagers. The reported male-to female ratio is approximately 1:1<ref name=":5">Orazi A, et al., (2017). | The median patient age at diagnosis is in the seventh or eighth decade of life<ref name=":0" /><ref name=":1">{{Cite journal|last=Jm|first=Hernández|last2=Mc|first2=del Cañizo|last3=A|first3=Cuneo|last4=Jl|first4=García|last5=Nc|first5=Gutiérrez|last6=M|first6=González|last7=G|first7=Castoldi|last8=Jf|first8=San Miguel|date=2000|title=Clinical, Hematological and Cytogenetic Characteristics of Atypical Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/10847463/|language=en|pmid=10847463}}</ref><ref name=":2">{{Cite journal|last=P|first=Martiat|last2=Jl|first2=Michaux|last3=J|first3=Rodhain|date=1991|title=Philadelphia-negative (Ph-) Chronic Myeloid Leukemia (CML): Comparison With Ph+ CML and Chronic Myelomonocytic Leukemia. The Groupe Français De Cytogénétique Hématologique|url=https://pubmed.ncbi.nlm.nih.gov/2070054/|language=en|pmid=2070054}}</ref>. Rare cases of aCML has also been reported in teenagers. The reported male-to female ratio is approximately 1:1<ref name=":5">Orazi A, et al., (2017). | ||
| Line 42: | Line 59: | ||
==Clinical Features== | ==Clinical Features== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |EXAMPLE Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | EXAMPLE B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | EXAMPLE Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | EXAMPLE Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |EXAMPLE Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | EXAMPLE Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 105: | Line 122: | ||
==Immunophenotype== | ==Immunophenotype== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 111: | Line 128: | ||
!Finding!!Marker | !Finding!!Marker | ||
|- | |- | ||
| − | |Positive (universal)||EXAMPLE CD1 | + | |Positive (universal)||<span class="blue-text">EXAMPLE:</span> CD1 |
|- | |- | ||
| − | |Positive (subset)||EXAMPLE CD2 | + | |Positive (subset)||<span class="blue-text">EXAMPLE:</span> CD2 |
|- | |- | ||
| − | |Negative (universal)||EXAMPLE CD3 | + | |Negative (universal)||<span class="blue-text">EXAMPLE:</span> CD3 |
|- | |- | ||
| − | |Negative (subset)||EXAMPLE CD4 | + | |Negative (subset)||<span class="blue-text">EXAMPLE:</span> CD4 |
|} | |} | ||
| Line 138: | Line 155: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 177: | Line 194: | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain / Loss / LOH== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 187: | Line 204: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |EXAMPLE Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 200: | Line 217: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |EXAMPLE Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 217: | Line 234: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 228: | Line 245: | ||
*Trisomy 8 and del(20q) most common. | *Trisomy 8 and del(20q) most common. | ||
*Abnormalities of chromosomes 13, 14, 17, 19 and 12 are also common <ref name=":1" /><ref name=":2" />. | *Abnormalities of chromosomes 13, 14, 17, 19 and 12 are also common <ref name=":1" /><ref name=":2" />. | ||
| − | *Isolated isochromosome 17q rare (most present with features of [[Chronic | + | *Isolated isochromosome 17q rare (most present with features of [[HAEM5:Chronic myelomonocytic leukaemia|CMML]] instead of aCML). |
</blockquote> | </blockquote> | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| − | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis'')</span> | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 243: | Line 260: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 249: | Line 266: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 261: | Line 278: | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV / INDEL)== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 271: | Line 288: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE: TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | EXAMPLE: | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | EXAMPLE: BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |EXAMPLE: TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |EXAMPLE: 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE: 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |EXAMPLE: IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |EXAMPLE: EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 322: | Line 339: | ||
|6%<ref name=":6" /> | |6%<ref name=":6" /> | ||
|} | |} | ||
| − | Mutations in ''TET2, ASXL1, EZH2'' and ''SRSF2'' (overlapping with [[Chronic | + | Mutations in ''TET2, ASXL1, EZH2'' and ''SRSF2'' (overlapping with [[HAEM5:Chronic myelomonocytic leukaemia|CMML]]) are not uncommon in aCML<ref>{{Cite journal|last=Faisal|first=Muhammad|last2=Stark|first2=Helge|last3=Büsche|first3=Guntram|last4=Schlue|first4=Jerome|last5=Teiken|first5=Kristin|last6=Kreipe|first6=Hans H.|last7=Lehmann|first7=Ulrich|last8=Bartels|first8=Stephan|date=2019|title=Comprehensive mutation profiling and mRNA expression analysis in atypical chronic myeloid leukemia in comparison with chronic myelomonocytic leukemia|url=http://doi.wiley.com/10.1002/cam4.1946|journal=Cancer Medicine|language=en|volume=8|issue=2|pages=742–750|doi=10.1002/cam4.1946|pmc=PMC6382710|pmid=30635983}}</ref>. |
</blockquote> | </blockquote> | ||
| Line 331: | Line 348: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table.'')</span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |EXAMPLE: BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |EXAMPLE: MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |EXAMPLE: Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |EXAMPLE: CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |EXAMPLE: Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |EXAMPLE: Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |EXAMPLE: KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |EXAMPLE: Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |EXAMPLE: Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
| Line 383: | Line 400: | ||
==Links== | ==Links== | ||
| − | Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page <span style="color:#0070C0">(''Instructions: | + | Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page <span style="color:#0070C0">(''Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "<nowiki>http://www</nowiki>." portion.'')</span> |
==References== | ==References== | ||
Latest revision as of 17:17, 6 September 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Atypical Chronic Myeloid Leukemia (aCML), BCR-ABL1 Negative.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Linsheng Zhang, MD, PhD
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Myelodysplastic/myeloproliferative neoplasms |
| Type | N/A |
| Subtype(s) | Myelodysplastic/myeloproliferative neoplasm with neutrophilia |
Definition / Description of Disease
Atypical chronic myeloid leukemia, BCR-ABL1-negative (aCML) presents with myelodysplastic as well as myeloproliferative features at the time of disease onset[1][2]. The characteristic finding is blood leukocytosis with increase of dysplastic neutrophils and immature granulocytes. Bone marrow shows multilineage dysplasia.
- Before the diagnosis of aCML can be rendered, myeloid neoplasms with well defined genetic abnormalities such as BCR-ABL1 fusion; rearrangement of PDGFRA, PDGFRB, FGFR1, or PCM1-JAK2 must be ruled out.
- Myeloid neoplasm with features of aCML developed after chemo or radiation therapy is classified as therapy-related myeloid neoplasm.
Synonyms / Terminology
Atypical chronic myeloid leukemia, Philadelphia chromosome-negative (Ph-)
Atypical chronic myeloid leukemia, BCR-ABL1-negative
Epidemiology / Prevalence
The incidence of aCML is low; it is estimated that there are 1-2 aCML cases for every 100 cases of BCR-ABL1-positive chronic myeloid leukemia[3][4].
The median patient age at diagnosis is in the seventh or eighth decade of life[4][5][6]. Rare cases of aCML has also been reported in teenagers. The reported male-to female ratio is approximately 1:1[1].
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.The primary symptoms are related to splenomegaly and blood cell changes like anemia and/or thrombocytopenia[5][7][6].
Sites of Involvement
- Blood and bone marrow are always involved.
- Involvement of spleen and liver is common.
Morphologic Features
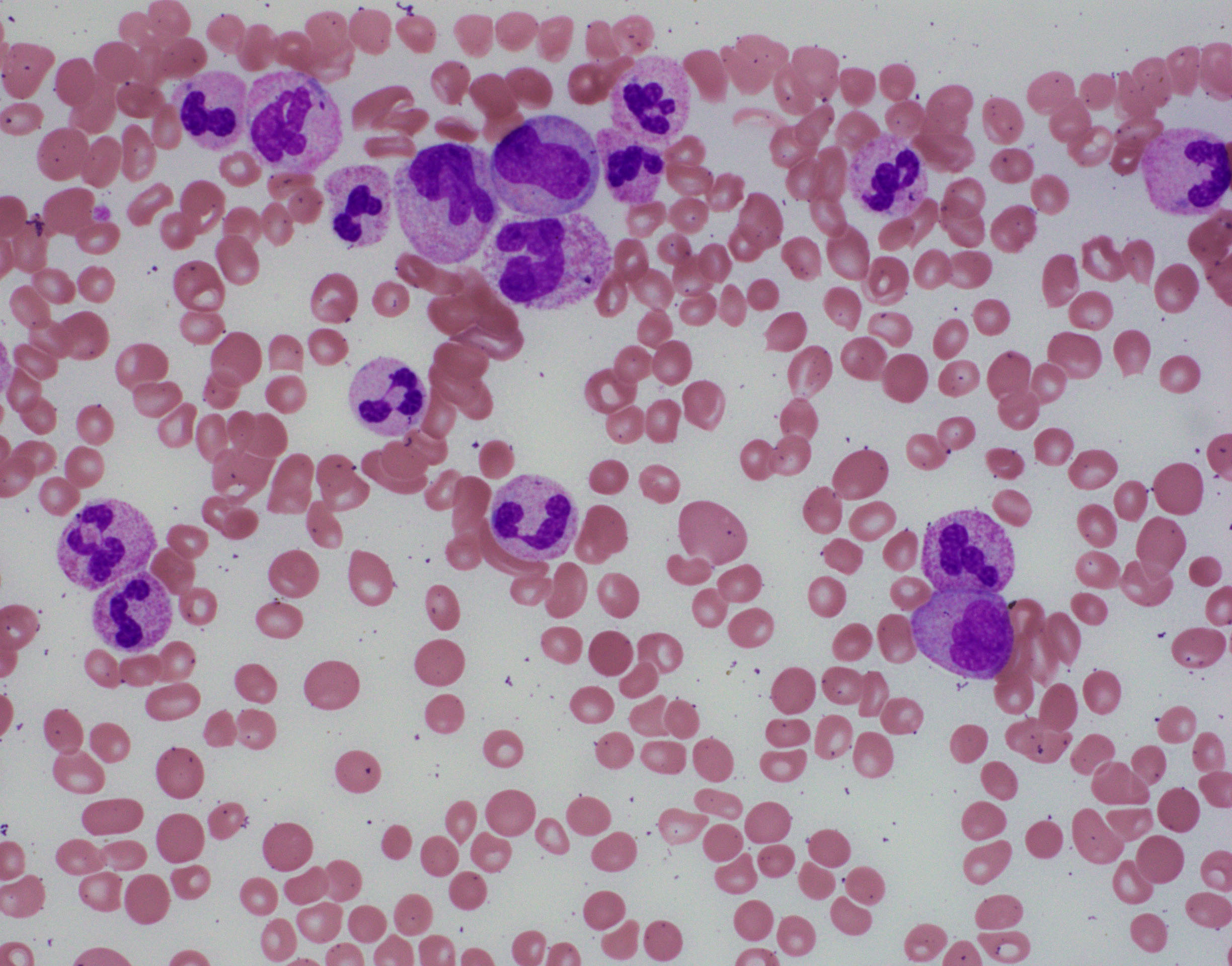
BLOOD:
- The white blood cell count (WBC) ≥13 x 109/L, cases >300 x 109/L have been reported.
- Immature granulocytes, including promyelocytes, myelocytes, and metamyelocytes ≥10% of the leukocytes.
- Monocytes <10%.
- Mild basophilia.
- Dysplastic features observed in granulocytes:
- Acquired Pelger-Huët anomaly
- Other nuclear abnormalities (hypersegmentation with abnormally clumped nuclear chromatin or bizarrely segmented nuclei)
- Multiple nuclear projections
- Abnormal cytoplasmic granularity (usually hypogranularity).
- The syndrome of abnormal chromatin clumping is considered to be a morphologic variant of aCML[8].
- Neutrophil nuclei show large well-separated blocks of heterochromatin and euchromatin
- Anemia and thrombocytopenia are common.
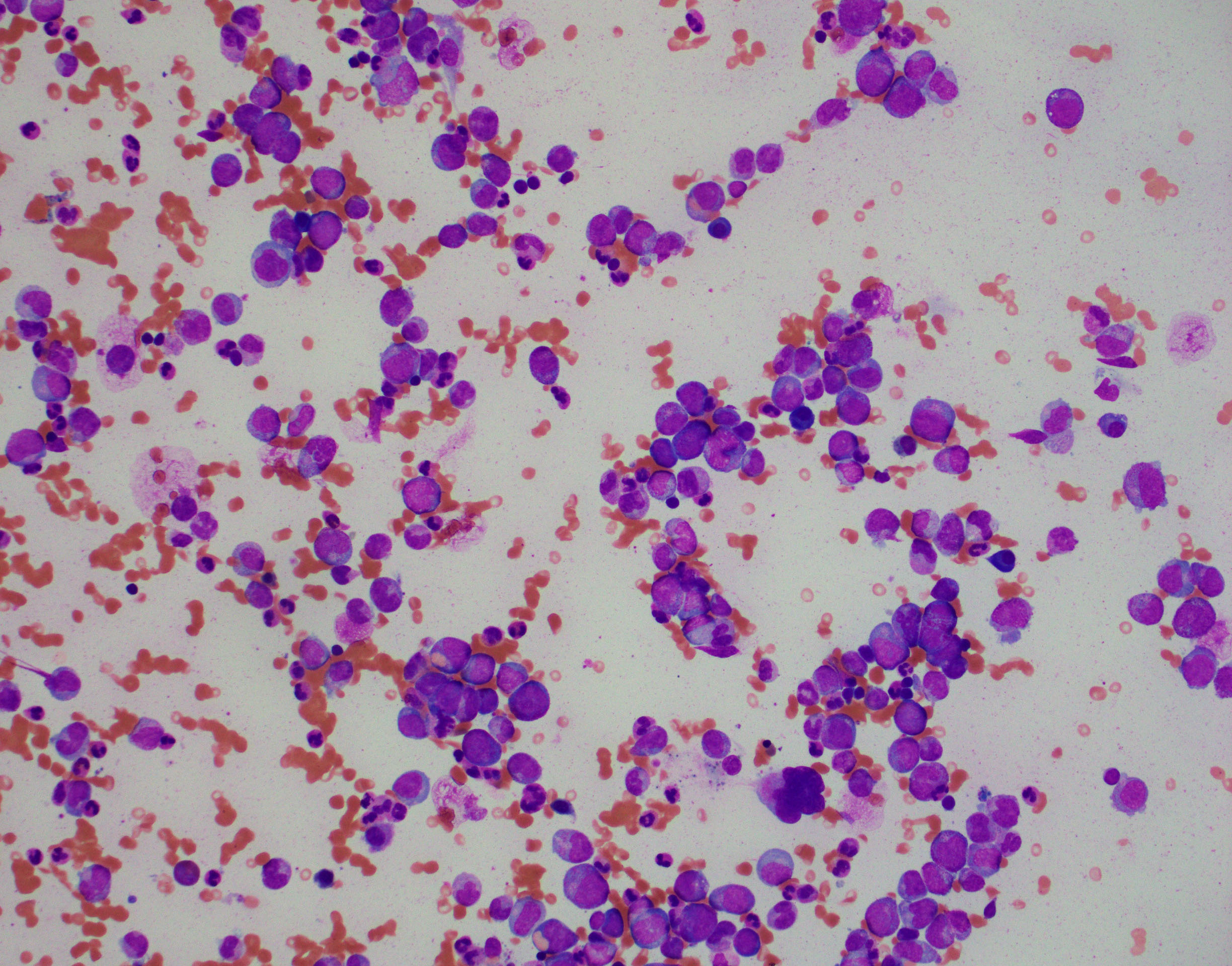
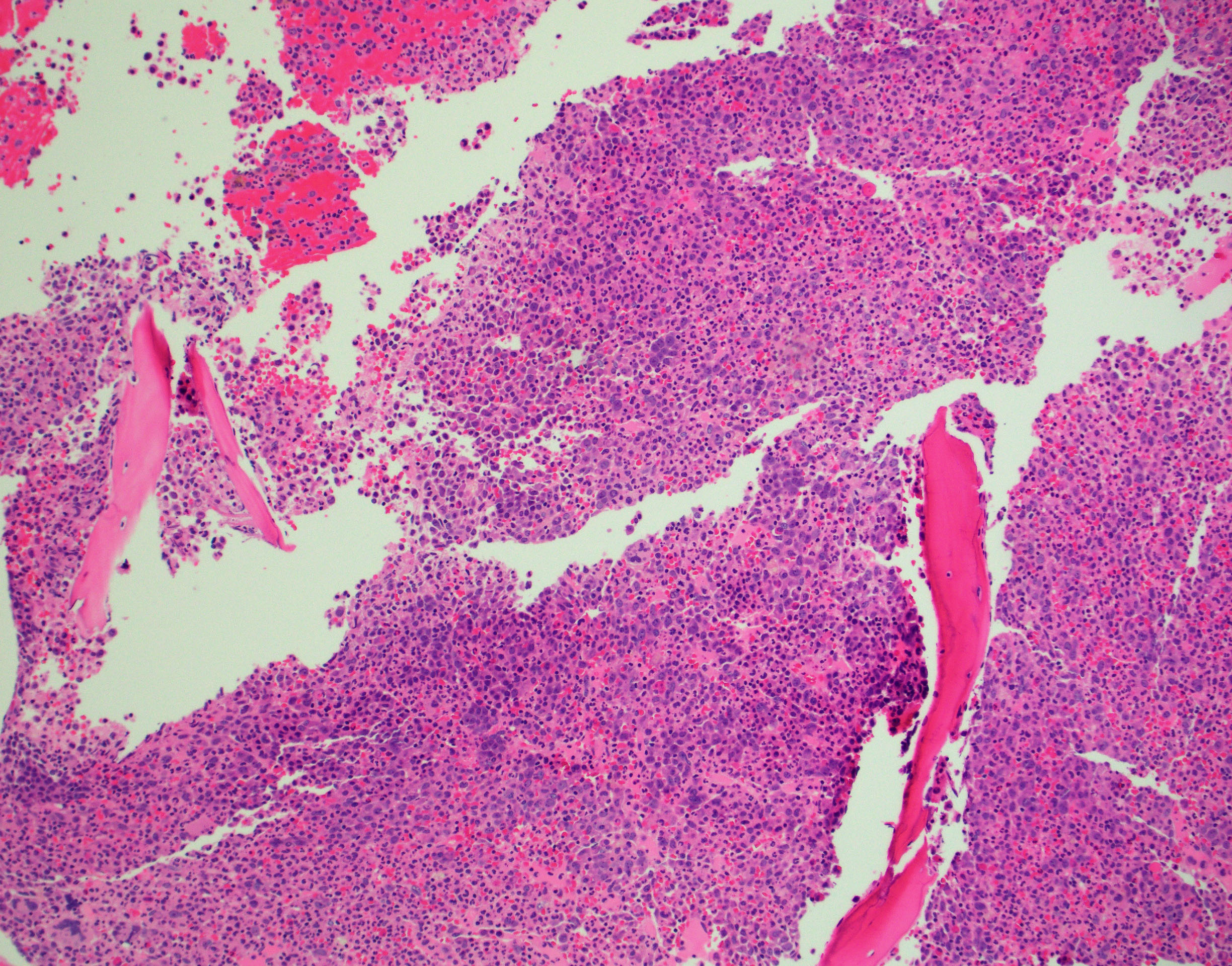
BONE MARROW:
- Hypercellular with markedly increased myeloid-to-erythroid ratio (usually >10:1)
- Features of trilineage dysplasia
- Dyserythropoiesis is present in about 40% of cases.
- Dysgranulopoiesis with the changes in the neutrophil lineage similar to those in the blood.
- Megakaryocytes quantitatively variable, usually normal or increased, frequently with dysmegakaryopoiesis, including micromegakaryocytes and small megakaryocytes with hypolobated nuclei.
- Mild fibrosis seen in some cases; may be more conspicuous with disease progression.
By definition, blasts <20% of blood leukocytes (usually <5%) and <20% of bone marrow nucleated cells.
Immunophenotype
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Finding | Marker |
|---|---|
| Positive (universal) | EXAMPLE: CD1 |
| Positive (subset) | EXAMPLE: CD2 |
| Negative (universal) | EXAMPLE: CD3 |
| Negative (subset) | EXAMPLE: CD4 |
editv4:ImmunophenotypeThe content below was from the old template. Please incorporate above.No specific immunophenotypic characteristics (flow cytometric analysis of blood and bone marrow usually report as normal, except mildly increased myeloblasts in some cases).
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.None.
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
Genetic abnormalities and mutations in related genes provide evidence of a clonal process to support a neoplastic process. However, they are not specific for aCML.
The prognostic value of molecular/genetic abnormalities are not well studied yet.
Approximately 20-40% of aCML evolves to acute myeloid leukemia[4]; most of the remaining patients die of marrow failure[7][9].
Predictors of worse prognosis:
- Age > 65 years
- Female sex
- White blood cell count > 50 x 109/L, thrombocytopenia and hemoglobin level < 10 g/dL[9].
- Increased blasts in blood and/or bone marrow.
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.Karyotypic abnormalities are reported in as many as 80% of cases.
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.None.
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.
Gene Mutation Oncogene/Tumor Suppressor/Other Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) Prevalence SETBP1 D868N, S869N, G870S, I871T and D880N[10] Other: conferring self-renewal capability to myeloid progenitors[11] GOF 25-38%[12][13] Other Mutations
Gene/Mutation Prevalence[4] RAS (KRAS/NRAS) 35% ETNK1: H243Y and N244S 8.8%[14] JAK2: V617F 7% CSF3R <10% CALR <10% APC2 6%[13] Mutations in TET2, ASXL1, EZH2 and SRSF2 (overlapping with CMML) are not uncommon in aCML[15].
Epigenomic Alterations
Not well studied yet.
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.Frequently involved:
- Cell proliferation and survival (SETBP1)
Less commonly involved:
- JAK/STAT pathway (JAK2)
- PI3K/AKT and MAPK/ERK (CSF3R)
Genetic Diagnostic Testing Methods
Blood cell counts and blood smear review (differential cell counts and dysplastic features).
Bone marrow aspirate smear review and biopsy histopathology (abnormal maturation pattern, blast percentage, etc.).
Molecular/genetic studies are of critical importance to establish clonal evidence and rule out other entities with well defined genetic abnormalities.
- Chromosome analysis
- Fluorescence in situ hybridization (FISH) to rule out PDGFRA, PDGFRB or FGFR1 rearrangement, BCR-ABL1 and PCM1-JAK2 fusion.
- Next generation sequencing-based mutation profiling to detect mutations supporting a clonal process.
Familial Forms
None
Additional Information
None
Links
Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page (Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "http://www." portion.)
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 Orazi A, et al., (2017). Atypical chronic myeloid leukaemia, BCR-ABL1-negative, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p87-89.
- ↑ Czader, M and Orazi, A. Chapter 45. Myelodysplastic/Myeloproliferative Neoplasms and Related Diseases. In Orazi A, Foucar K, Knowles DM. eds. Knowles Neoplastic Hematopathology. Riverwoods, IL: Wolters Kluwer Health; 2013:1148-1150.
- ↑ A, Orazi; et al. (2008). "The Myelodysplastic/Myeloproliferative Neoplasms: Myeloproliferative Diseases With Dysplastic Features". PMID 18480833.
- ↑ 4.0 4.1 4.2 4.3 Sa, Wang; et al. (2014). "Atypical Chronic Myeloid Leukemia Is Clinically Distinct From Unclassifiable Myelodysplastic/Myeloproliferative Neoplasms". doi:10.1182/blood-2014-02-553800. PMC 4067498. PMID 24627528.CS1 maint: PMC format (link)
- ↑ 5.0 5.1 5.2 Jm, Hernández; et al. (2000). "Clinical, Hematological and Cytogenetic Characteristics of Atypical Chronic Myeloid Leukemia". PMID 10847463.
- ↑ 6.0 6.1 6.2 P, Martiat; et al. (1991). "Philadelphia-negative (Ph-) Chronic Myeloid Leukemia (CML): Comparison With Ph+ CML and Chronic Myelomonocytic Leukemia. The Groupe Français De Cytogénétique Hématologique". PMID 2070054.
- ↑ 7.0 7.1 R, Kurzrock; et al. (2001). "BCR Rearrangement-Negative Chronic Myelogenous Leukemia Revisited". PMID 11387365.
- ↑ Brizard, A.; et al. (1989). "THREE CASES OF MYELODYSPLASTIC-MYELOPROLIFERATIVE DISORDER WITH ABNORMAL CHROMATIN CLUMPING IN GRANULOCYTES". British Journal of Haematology. 72 (2): 294–295. doi:10.1111/j.1365-2141.1989.tb07703.x. ISSN 0007-1048.
- ↑ 9.0 9.1 M, Breccia; et al. (2006). "Identification of Risk Factors in Atypical Chronic Myeloid Leukemia". PMID 17043019.
- ↑ R, Acuna-Hidalgo; et al. (2017). "Overlapping SETBP1 Gain-Of-Function Mutations in Schinzel-Giedion Syndrome and Hematologic Malignancies". doi:10.1371/journal.pgen.1006683. PMC 5386295. PMID 28346496.CS1 maint: PMC format (link)
- ↑ Oakley, Kevin; et al. (2012). "Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10". Blood. 119 (25): 6099–6108. doi:10.1182/blood-2011-10-388710. ISSN 0006-4971. PMC 3383018. PMID 22566606.CS1 maint: PMC format (link)
- ↑ Mughal, T. I.; et al. (2015). "An International MDS/MPN Working Group's perspective and recommendations on molecular pathogenesis, diagnosis and clinical characterization of myelodysplastic/myeloproliferative neoplasms". Haematologica. 100 (9): 1117–1130. doi:10.3324/haematol.2014.114660. ISSN 0390-6078. PMC 4800699. PMID 26341525.CS1 maint: PMC format (link)
- ↑ 13.0 13.1 Palomo, Laura; et al. (2020). "Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms". Blood. doi:10.1182/blood.2019004229. ISSN 0006-4971.
- ↑ Gambacorti-Passerini, Carlo B.; et al. (2015). "Recurrent ETNK1 mutations in atypical chronic myeloid leukemia". Blood. 125 (3): 499–503. doi:10.1182/blood-2014-06-579466. ISSN 0006-4971.
- ↑ Faisal, Muhammad; et al. (2019). "Comprehensive mutation profiling and mRNA expression analysis in atypical chronic myeloid leukemia in comparison with chronic myelomonocytic leukemia". Cancer Medicine. 8 (2): 742–750. doi:10.1002/cam4.1946. PMC 6382710. PMID 30635983.CS1 maint: PMC format (link)
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Myelodysplastic/myeloproliferative neoplasm with neutrophilia”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 09/6/2024, https://ccga.io/index.php/HAEM5:Myelodysplastic/myeloproliferative_neoplasm_with_neutrophilia.