HAEM4Backup:Myeloid Leukemia Associated with Down Syndrome
Primary Author(s)*
Linda D Cooley, MD, MBA, Children's Mercy Hospital, Kansas City, MO
Cancer Category/Type
Myeloid Proliferations Associated with Down Syndrome
Cancer Sub-Classification / Subtype
Myeloid leukemia associated with Down syndrome
Definition / Description of Disease
This is a distinct entity in the World Health Organization (WHO) classification system within the section of Myeloid Proliferations Associated with Down Syndrome[1].
In Down Syndrome (DS), most cases of AML are acute megakaryoblastic leukemia (AMKL). AMKL accounts for >50% of all cases of acute leukemia in DS beyond the neonatal period. AML often follows a myelodysplastic syndrome (MDS)-like phase. In DS individuals, there is no biological difference between MDS and overt AML[1].
Acute megakaryoblastic leukemia (AMKL) is a subtype of acute myeloid leukemia (AML) characterized by abnormal megakaryoblasts that express platelet-specific surface glycoprotein. Bone marrow biopsy frequently demonstrates extensive myelofibrosis, often making aspiration in these patients difficult. AMKL comprises between 4% and 15% of pediatric AML patients. It is divided into 2 major subgroups: AMKL in patients with Down syndrome (DS-AMKL) and AMKL in patients without DS (non-DS-AMKL). AMKL is the most frequent type of AML in children with DS, and the incidence in these patients is 500-fold higher than in the general population.
DS-AMKL is biologically and clinically distinct from non-DS-AMKL. Pediatric non-DS-AMKL (i.e. AMKL NOT associated with Down syndrome) occurs in a heterogenous group of patients, a significant proportion of whom carry chimeric oncogenes including RBM15-MKL1, which represents the distinct entity Acute Myeloid Leukemia (AML) Megakaryoblastic with t(1;22)(p13.3;q13.1);RBM15-MKL1, CBFA2T3-GLIS2, NUP98-KDM5A, and KMT2A(MLL) rearrangements, which are not found in DS-AMKL[2].
Synonyms / Terminology
Acute myeloid leukemia associated with Down syndrome
Epidemiology / Prevalence
Children with DS have a markedly increased risk (~150-fold) of developing AML compared with children without DS.
The majority of children with myeloid leukemia associated with Down syndrome (ML-DS) are <5 years of age. Approximately 1-2% of DS children develop AML during the first 5 years of life. Children with DS account for ~20% of all pediatric patients with AML/MDS. Myeloid leukemia associated with DS occurs in 20-30% of children with a history of transient abnormal myelopoiesis (TAM) and the leukemia usually occurs 1-3 years after TAM[1].
Clinical Features
ML-DS presents at a median age of 1-1.8 years and is rare after the age of 4 years. Most case of ML-DS have a history consistent with a preceding TAM in the neonatal period. If there is no history, most likely the appropriate diagnostic tests were not done at birth; GATA1 mutations are found on neonatal bloodspots in patients with ML-DS. The presentation of ML-DS is often indolent with myelodysplasia and progressive pancytopenia, particularly thrombocytopenia and leukopenia, with a low percentage of circulating blasts for months before development of ML-DS. A bone marrow aspirate may be needed to make the diagnosis, however, due to marrow fibrosis, a bone marrow biopsy may be necessary[2][3].
Sites of Involvement
Blood and bone marrow are the principle sites of involvement. Extramedullary involvement, mainly of the spleen and liver, is almost always present as well[1].
Morphologic Features
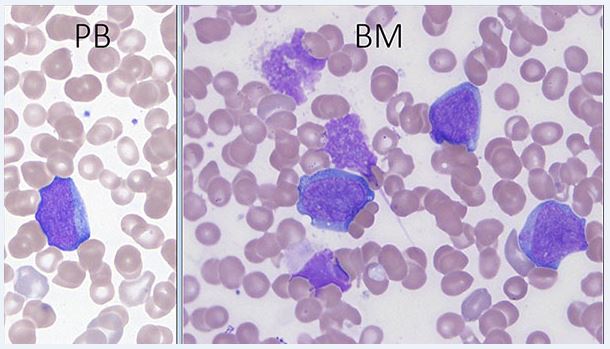
In the preleukemic phase, the disease has features of refractory cytopenia of childhood and lacks a significant increase in blasts. Erythroid cells are macrocytic. Dysplastic features may be more pronounced than in primary refractory cytopenia. In AML, blasts and occasionally erythroid precursors are usually present in the peripheral blood. Erythrocytes often show anisopoikilocytosis, and dacryocytes can be seen. Platelets are usually decreased and giant platelets may be present. The bone marrow aspirate blast morphology shows round to slightly irregular nuclei and a moderate amount of basophilic cytoplasm. Cytoplasmic blebs may be present. The cytoplasm of blasts may contain coarse granules; the granules are generally MPO-negative. Erythroid precursors often show megaloblastic changes and dsyplastic forms, including binucleated or trinucleated cells and nuclear fragments. Dysgranulopoiesis may be present. The bone core may show dense reticulin, making marrow aspiration difficult. Megakaryocytes may be markedly increased with dysplastic small forms and micromegakaryocytes[1].
Immunophenotype
In most cases, the leukemic blasts are positive for KIT (CD117), CD13, CD33, CD11b, CD7, CD4, CD42, CD110 (TPOR), IL3R, CD36, CD41, CD61, and CD71. The blasts are negative for MPO, CD15, CD14, and glycophorin A. CD34 is negative in 50% of cases, and ~30% of cases are negative for CD56 and CD41[1].
Chromosomal Rearrangements (Gene Fusions)
Chromosomal rearrangements and gene fusions are not characteristic of Myeloid leukemia associated with Down syndrome[4].
Characteristic Chromosomal Aberrations / Patterns
The most frequent chromosomal alterations associated with ML-DS are dup(1q), del(6q), del(7p), dup(7q), +8, +11, del(16q) and +21. The karyotypic patterns of ML-DS are different from those observed in AML of children without DS, e.g. translocations t(8;21), t(15;17), t(9;11), inversion inv(16), as well as AMKL associated translocations t(1;22) and t(1;3)[4].
Genomic Gain/Loss/LOH
See chromosomal aberrations
Gene Mutations (SNV/INDEL)
| Gene | Mutation | Oncogene/Tumor Suppressor/Other | Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) | Prevalence (COSMIC/TCGA/Other) |
|---|---|---|---|---|
| GATA1 | Most mutations insert a premature termination codon either by introducing a stop codon or frameshift. Mutations affecting the splice site at GATA1 exon 2 exon/intron boundary are next most frequent. | zinc finger DNA-binding transcription factor that plays a critical role in the normal development of hematopoietic cell lineages | N-terminally truncating somatic mutation | 100% |
Other Mutations
| Type | Gene/Region/Other |
|---|---|
| Secondary Mutations | Cohesin (STAG2, RAD21, SMC3, SMC1A, NIPBL)
Epigenetic regulators (EZH2, SUZ12) Signaling molecules (JAK1, JAK2, JAK3, MPL, KRAS, NRAS, SH2B3) |
| Mutually Exclusive | EZH2 and SUZ12 |
Epigenomics (Methylation)
EZH2, the catalytic subunit of the Polycomb repressive complex 2 (PRC2) is the most frequently targeted epigenetic regulator in DS-AMKL. SUZ12 and EZH2 are found in ~35% of cases; PRC2 often combined with alterations in CTCF or cohesin. In erythroid cells, PRC2 is involved in epigenetic silencing of a subset of GATA1-repressed genes. Disruption of the repression may therefore enhance the self-renewal of cells, contributing to the differentiation block provided by the truncated GATA1 protein[2].
Genes and Main Pathways Involved
The cellular and molecular events involved in initiation and evolution of TAM and ML-DS can be understood as a three-step model which requires the presence within a fetal liver-derived haematopoietic stem or progenitor cell of (i) trisomy 21, (ii) an acquired GATA1 mutation, and (iii) at least one additional oncogenic mutation.
Trisomy 21, the initial event, causes an increase in numbers of megakaryocyte-erythroid progenitors (MEP) and an increase in the hematopoietic stem cell (HSC) compartment. Megakaryocyte differentiation is impaired and platelet counts are reduced both in fetal blood and in neonates with DS. Studies have implicated increased expression of various genes on chromosome 21, in particular ERG and DYRK1a, as important mediators of the abnormal megakaryopoiesis. Recent data suggest that trisomy of RUNX1, ETS2, and ERG might be sufficient, in combination with mutant GATA1, to explain the hematopoietic abnormalities in human fetal liver and TAM cells. Trisomy 21 causes genome-wide changes in gene expression directly or indirectly affecting multiple genes on most chromosomes.
N-terminal truncating GATA1 mutations can be detected in all cases. These acquired mutations disappear when TAM (or ML-DS) enter remission. GATA1 mutations are present in all cases of TAM or ML-DS and they are present in 25-30% of all neonates with Down syndrome. GATA1 mutations are necessary for the development of TAM/ML-DS. Multiple GATA1 mutant clones are present in up to 25% of neonates with Down syndrome. The majority (~97%) of GATA1 mutations (insertions, deletions and point mutations) are found in exon 2 and the remainder in exon 3.1. Mutations lead to expression of a truncated GATA1s protein. The type of GATA1 mutation does not predict which patients with TAM will later progress to ML-DS. GATA1 is a regulator of normal megakaryocyte and erythroid differentiation.
The presence of an N-terminal truncating mutation in GATA1 is necessary, but insufficient, for development of ML-DS. Mutations in the key cohesin component genes (RAD21, STAG2, SMC3, SMC1A, CTCF) and in epigenetic regulators such as EZH2 and KANSL1 have been shown to occur with high frequency. These genes encode proteins important for transcription regulation. RAS pathway genes (NRAS, KRAS, CBL, PTPN11, NF1) are seen in a smaller proportion of patients. The fetal liver hematopoietic microenvironment may contribute both to expansion and/or maintenance of the mutant GATA1 clone in TAM. The natural history and clinical features of TAM show this to be a fetal liver disease[2][3].
Diagnostic Testing Methods
Routine chromosome analysis and microarray analysis should be done for all new diagnosis leukemias. If DS is suspected, but not yet confirmed, a PHA-stimulated blood chromosome study should be done to confirm the diagnosis of constitutional +21 and determine if mosaicism is present. Microarray analysis at diagnosis will determine the presence of clonal aberrations that may be undetected by routine chromosome analysis, as well as uncover any additional constitutional aberrations that may be present.
GATA1 gene sequencing analysis using Sanger or NGS methodologies may be done; however, as all DS patients with TAM or myeloid leukemia have been shown to have GATA1 mutation, it may be determined to be an optional test. WES may reveal acquired mutations in genes which cooperate with the GATA1 mutations to transform TAM to ML-DS.
Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications)
Children with ML-DS have better outcomes compared to children without Down syndrome with long-term survival of 74-91%. Treatment failure in children with Down syndrome can be due to chemotherapy-related toxicity, thus most protocols now use reduced dose of anthracycline that is less toxic and has similar efficacy.
The outlook for children with ML-DS who relapse is very poor with a 3-year overall survival rate of 20-25%. There appears to be no role for allogeneic stem cell transplant (SCT) in first-line therapy for ML-DS due to efficacy of chemotherapy and the ongoing high rate of toxicity associated with SCT. The role of SCT as salvage therapy is unclear as the 3-year probability of overall survival of only 19% due to the high rate of relapse and transplant-related mortality[2].
Familial Forms
not applicable
Other Information
Also see: Transient Abnormal Myelopoiesis (TAM) Associated with Down Syndrome
Links
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5031718/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5594429/
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Arber DA, et al., (2017). Myeloid proliferations associated with Down syndrome, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p170-171.
- ↑ 2.0 2.1 2.2 2.3 2.4 Gruber, Tanja A.; et al. (2015). "The biology of pediatric acute megakaryoblastic leukemia". Blood. 126 (8): 943–949. doi:10.1182/blood-2015-05-567859. ISSN 1528-0020. PMC 4551356. PMID 26186939.
- ↑ 3.0 3.1 Bhatnagar, Neha; et al. (2016). "Transient Abnormal Myelopoiesis and AML in Down Syndrome: an Update". Current Hematologic Malignancy Reports. 11 (5): 333–341. doi:10.1007/s11899-016-0338-x. ISSN 1558-822X. PMC 5031718. PMID 27510823.
- ↑ 4.0 4.1 de Souza, Daiane Correa; et al. (2017). "A unique set of complex chromosomal abnormalities in an infant with myeloid leukemia associated with Down syndrome". Molecular Cytogenetics. 10: 35. doi:10.1186/s13039-017-0335-3. ISSN 1755-8166. PMC 5594429. PMID 28912835.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.