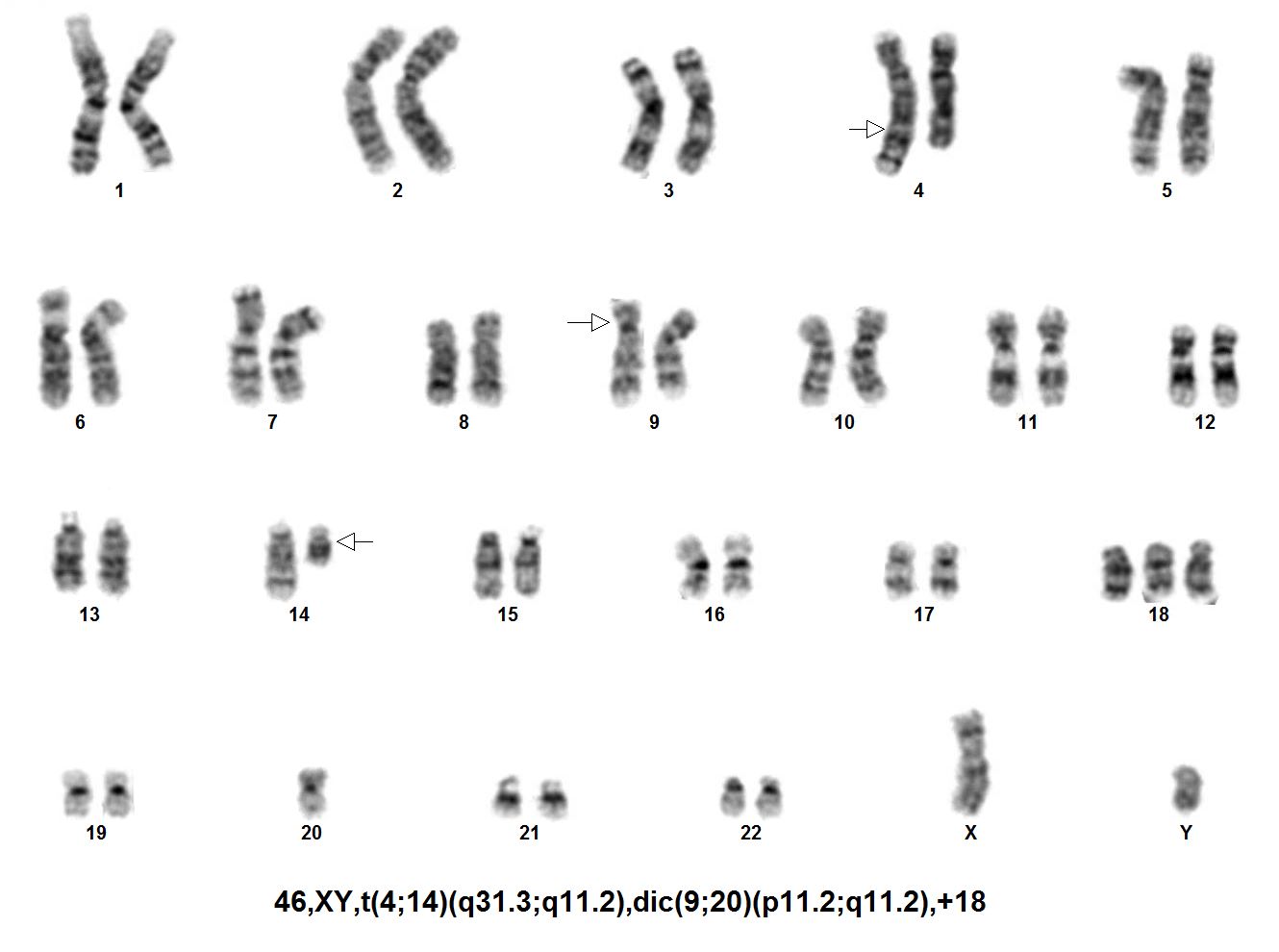
HAEM4Backup:B-Lymphoblastic Leukemia/Lymphoma with dic(9;20)(p13;q11)
| This page is under construction |
Primary Author(s)*
Alaa Koleilat, Ph.D., Mayo Clinic and Xinjie Xu, Ph.D., Mayo Clinic
Cancer Category/Type
B-lymphoblastic leukemia/lymphoma
Cancer Sub-Classification / Subtype
B-lymphoblastic leukemia/lymphoma with dic(9;20)(p13;q11)
Definition / Description of Disease
Neoplasm of B-cell lineage precursor lymphoblasts where the blasts contain alterations in transcription factor PAX5.
Synonyms / Terminology
- B- Lymphoblastic Leukemia (B-ALL) is also known as Precursor B-Cell Lymphoblastic Leukemia.
- dic(9;20)(p13;q11) is sometimes referred to as dic(9;20).
Epidemiology / Prevalence
- Occurs in approximately 2% of pediatric patients with B-ALL and 0.5% of adult precursor B-ALL[1]
- The male to female ratio is 1 to 2.[2]
- The median age of diagnosis is 4 years.[3][2]
Clinical Features
- The median leucocyte count is 20–30 × 109/l.[4]
- The event-free survival (EFS) at 5 years is 62% and the overall survival (OS) up to 5 years is 82%.
Sites of Involvement
Bone marrow, Peripheral Blood
Morphologic Features
Put your text here
Immunophenotype
Put your text here and/or fill in the table
| Finding | Marker |
|---|---|
| Positive (universal) | HLA-DR, CD10, CD19, CD20, CD22 |
| Positive (subset) | EXAMPLE CD2 |
| Negative (universal) | T cell and myeloid markers |
| Negative (subset) | EXAMPLE CD4 |
Chromosomal Rearrangements (Gene Fusions)
There are no fusion partners that have been reported as a result of dic(9;20)(p13;q11); however, breakpoints that occur within the PAX5 gene have resulted in fusion sequences with regions of chr. 20 including ASXL1, C20ORF112 and KIF3B.[5]
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence |
|---|---|---|---|
| EXAMPLE t(9;22)(q34;q11.2) | EXAMPLE 3'ABL1 / 5'BCR | EXAMPLE der(22) | EXAMPLE 5% |
| EXAMPLE t(8;21)(q22;q22) | EXAMPLE 5'RUNX1 / 3'RUNXT1 | EXAMPLE der(8) | EXAMPLE 5% |
Characteristic Chromosomal Aberrations / Patterns
- In 40% of ALL cases, the dic(9;20)(p13;q11) can be found as a sole chromosomal aberration and the remaining 60% it occurs with additional chromosomal aberrations.[2]
- The dic(9;20)(p13;q11) easily escapes detection by traditional cytogenetic approaches including karyotyping which rely heavily on the identification of monosomy 20 as a marker for the abnormality; however, many cases with dic(9;20)(p13;q11) have two normal chromosome 20 homologs in addition to the rearranged one.[6][7]
- Results in a net loss of 9p and 20q
- Causes alterations of the transcription factor PAX5. The proposed driver of this leukemia subgroup has been deregulation of PAX5 expression due to full or partial deletions.
- There is evidence of clustering of breakpoints within 9p13.2 and 20q11.2.
- Deletions involving chromosome 13q are common genetic changes in ALL with dicentric (9;20)[9] as well as deletion of the second copy of 9p21.3 (CDKN2A/B).
- BCR-ABL1 and ETV6-RUNX1 fusion genes can co-occur with dic(9;20)(p13;q11).[10]
- Most cases have chromosome numbers between 44 and 50.[2]
- Although dic(9;20)(p13;q11) is mostly observed in B-ALL, it has been observed in a patient with T-ALL.[7]
Genomic Gain/Loss/LOH
| Chromosome Number | Gain/Loss/Amp/LOH | Region |
|---|---|---|
| 9 | Loss | 9p13-9pter (including CDKN2A/B) |
| 20 | Loss | 20q11-20qter (including ASXL1 and NOL4L) |
Gene Mutations (SNV/INDEL)
Put your text here and/or fill in the tables
| Gene | Mutation | Oncogene/Tumor Suppressor/Other | Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) | Prevalence (COSMIC/TCGA/Other) |
|---|---|---|---|---|
| EXAMPLE TP53 | EXAMPLE R273H | EXAMPLE Tumor Suppressor | EXAMPLE LOF | EXAMPLE 20% |
Other Mutations
| Type | Gene/Region/Other |
|---|---|
| Concomitant Mutations | EXAMPLE IDH1 R123H |
| Secondary Mutations | EXAMPLE Trisomy 7 |
| Mutually Exclusive | EXAMPLE EGFR Amplification |
Epigenomics (Methylation)
Put your text here
Genes and Main Pathways Involved
PAX5- was shown to be altered in 81% of ALL cases with a 9p abnormality.[11]
Diagnostic Testing Methods
- Conventional chromosome analysis (although can be difficult to detect with poor chromosome morphology)
- FISH analysis, interphase and metaphase if abnormal metaphases are observed (PAX5 BAP, centromeric probes for chromosomes 9 and 20 or subtelomeric probes for 20pter/20qter)
- Chromosomal microarray (CMA)
- Quantitative PCR can also be used to confirm disruption of PAX5 and downstream target genes
- Next-generation sequencing (NGS) based fusion panels can detect the PAX5 fusion.
Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications)
Diagnosis: The dicentric (9;20)(p13;q11) is more common in the pediatric population. Patients typically have a high WBC count and high frequency of CNS disease or extra-medullary leukemia.
Prognosis: The outcome for dic(9;20)(p13;q11) -positive pediatric ALL is not favorable as relapses are quite common, however post-relapse treatment of many patients is often successful.[2]
Therapeutic Implications: Most patients undergo non-standard risk treatment.[8]
Familial Forms
Put your text here
Other Information
There are over 200 dic(9;20) ALL cases in the Mitelman database.[12]
Links
References
(use "Cite" icon at top of page)
- ↑ Mitelman, F.; et al. (1997-04). "A breakpoint map of recurrent chromosomal rearrangements in human neoplasia". Nature Genetics. 15 Spec No: 417–474. doi:10.1038/ng0497supp-417. ISSN 1061-4036. PMID 9140409. Check date values in:
|date=(help) - ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Forestier, Erik; et al. (2008-02). "Clinical and cytogenetic features of pediatric dic(9;20)(p13.2;q11.2)-positive B-cell precursor acute lymphoblastic leukemias: a Nordic series of 24 cases and review of the literature". Genes, Chromosomes & Cancer. 47 (2): 149–158. doi:10.1002/gcc.20517. ISSN 1098-2264. PMID 17990329. Check date values in:
|date=(help) - ↑ Akkari, Yassmine M. N.; et al. (2020-05). "Evidence-based review of genomic aberrations in B-lymphoblastic leukemia/lymphoma: Report from the cancer genomics consortium working group for lymphoblastic leukemia". Cancer Genetics. 243: 52–72. doi:10.1016/j.cancergen.2020.03.001. ISSN 2210-7762. PMID 32302940 Check
|pmid=value (help). Check date values in:|date=(help) - ↑ Johansson, Bertil; et al. (2004). "Clinical and biological importance of cytogenetic abnormalities in childhood and adult acute lymphoblastic leukemia". Annals of Medicine. 36 (7): 492–503. doi:10.1080/07853890410018808. ISSN 0785-3890. PMID 15513300.
- ↑ An, Qian; et al. (2008-11-04). "Variable breakpoints target PAX5 in patients with dicentric chromosomes: a model for the basis of unbalanced translocations in cancer". Proceedings of the National Academy of Sciences of the United States of America. 105 (44): 17050–17054. doi:10.1073/pnas.0803494105. ISSN 1091-6490. PMC 2579376. PMID 18957548.
- ↑ Zachariadis, V.; et al. (2014-01). "Detecting dic(9;20)(p13.2;p11.2)-positive B-cell precursor acute lymphoblastic leukemia in a clinical setting using fluorescence in situ hybridization". Leukemia. 28 (1): 196–198. doi:10.1038/leu.2013.189. ISSN 1476-5551. PMID 23787394. Check date values in:
|date=(help) - ↑ 7.0 7.1 Clark, R.; et al. (2000-02). "Monosomy 20 as a pointer to dicentric (9;20) in acute lymphoblastic leukemia". Leukemia. 14 (2): 241–246. doi:10.1038/sj.leu.2401654. ISSN 0887-6924. PMID 10673740. Check date values in:
|date=(help) - ↑ 8.0 8.1 Zachariadis, V.; et al. (2011-04). "The frequency and prognostic impact of dic(9;20)(p13.2;q11.2) in childhood B-cell precursor acute lymphoblastic leukemia: results from the NOPHO ALL-2000 trial". Leukemia. 25 (4): 622–628. doi:10.1038/leu.2010.318. ISSN 1476-5551. PMID 21242996. Check date values in:
|date=(help) - ↑ Wafa, Abdulsamad; et al. (2020). "An acquired stable variant of a dicentric dic(9;20) and complex karyotype in a Syrian childhood B-acute lymphoblastic leukemia case". Molecular Cytogenetics. 13: 29. doi:10.1186/s13039-020-00499-x. ISSN 1755-8166. PMC 7350665 Check
|pmc=value (help). PMID 32670411 Check|pmid=value (help). - ↑ Gibbons, B. (1999). "dic(9;20)(p11–13;q11)". Atlas Genet Cytogenet Oncol Haematol. 3(3): 144.CS1 maint: display-authors (link)
- ↑ Coyaud, Etienne; et al. (2010-04-15). "Wide diversity of PAX5 alterations in B-ALL: a Groupe Francophone de Cytogenetique Hematologique study". Blood. 115 (15): 3089–3097. doi:10.1182/blood-2009-07-234229. ISSN 1528-0020. PMID 20160164.
- ↑ Mitelman F, Johansson B, Mertens F, editors. Mitelman database of chromosome aberrations and gene fusions in cancer (2019). http://cgap.nci.nih.gov/Chromosomes/Mitelman. Accessed 07.13.2021.
Book
- Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p129-171.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.