B-Lymphoblastic Leukemia/Lymphoma with TCF3 Rearrangements - Excluding t(1;19) and t(12;19)
editPREVIOUS EDITIONThis page from the 4th edition of Haematolymphoid Tumours is being updated. See 5th edition Table of Contents.
| This page is under construction |
Primary Author(s)*
Celeste Eno, PhD, Cedars-Sinai Medical Center
Cancer Category/Type
B-lymphoblastic leukemia/lymphoma (B-ALL/LBL) with TCF3 variant fusions
Cancer Sub-Classification / Subtype
TCF3::HLF Fusion t(17;19)(q22;p13.3)
Definition / Description of Disease
B-lymphoblastic leukemia/lymphoma (B-ALL/LBL) with TCF3::HLF harbors a translocation between TCF3 on chromosome 19 and HLF on chromosome 17. Variant TCF3 fusions are not associated with the WHO-classified entity, t(1;19), due to a difference in prognosis although the clinical features and immunoprofiles are reportedly similar.
Synonyms / Terminology
TCF3::HLF (E2A::HLF)
Epidemiology / Prevalence
t(17;19) is a rare translocation with an incidence of <1% in BCP-ALL and tends to present in pediatric and adolescents.[1][2][3]
Clinical Features
The presenting features are generally similar to those seen in patients with other ALL categories. Specific symptoms which have been noted to be associated with t(17;19) include coagulopathy and hypercalcemia.[4] Patients with TCF3::HLF Type I fusions have been suggested to be associated with disseminated intravascular coagulation. Patients with TCF3::HLF Type II fusions have been associated with hypercalcemia. It has been hypothesized that hypercalcemia in t(17;19) patients is partly parathyroid hormone-related protein (PTHrP) mediated.[4][1]
| Signs and Symptoms | B-symptoms (weight loss, fever, night sweats)
Coagulopathy[4] Type I: Disseminated intravascular coagulation[4][1] Type II: Hypercalcemia[4] |
| Laboratory Findings |
Sites of Involvement
Bone Marrow and Peripheral Blood
Morphologic Features
There are no unique morphological or cytochemical features that distinguish this entity from other types of ALL.
Immunophenotype
t(17;19) display an immunoprofile similar to t(1;19). Cases of t(1;19)-ALL with concomitant TCF3::PBX1 fusion invariably express a characteristic but uncommon profile of surface antigens. Blasts typically have a pre-B phenotype, with positivity for CD19, CD10, and cytoplasmic mu heavy chain. This diagnosis can be suspected even when cytoplasmic mu is not determined because these leukemias typically show strong expression of CD9 and lack CD34 or show very limited CD34 expression on only a minor subset of leukemic cells.[5][6]
| Finding | Marker |
|---|---|
| Positive (universal) | Cytoplasmic µ (cµ) heavy chain (pre-B phenotype); strong CD9; CD10; CD19 |
| Positive (subset) | CD22 |
| Negative (universal) | CD34; CD20 (Partial); CD21; Surface Immunoglobulin |
| Negative (subset) |
Chromosomal Rearrangements (Gene Fusions)
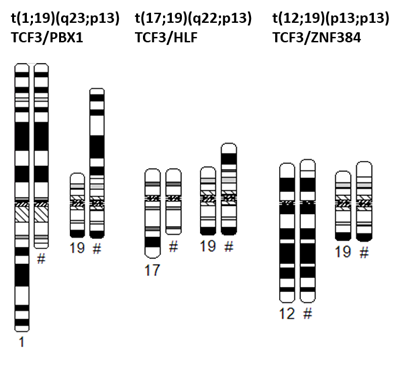
TCF3 encodes several transcription factors via alternative splicing. t (17;19) occurs in rare cases of ALL involving HLF (hepatic leukemia factor) on chromosome 17. The TCF3::HLF fusion gene encodes a chimeric transcription factor in which the transactivation domain of TCF3 links to the basic leucine zipper dimerization and DNA-binding domain of HLF.[8][9] There are two types of fusion: type 1 is generated by the fusion between exon 16 of TCF3 and exon 4 of HLF with an insertion of cryptic exon (joining region) maintaining an open-reading frame, whereas type 2 is generated by the fusion between exon 15 of TCF3 and exon 4 of HLF in the same reading frame.[2]
| Chromosomal Rearrangement | Genes in Fusion
(5’ or 3’ Segments) |
Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
| t(17;19)(q22;p13) | 5' TCF3 / 3' HLF | der(19) | <1% | Yes | Yes | No | There are two types of fusion: type 1 is generated by the fusion between exon 16 of TCF3 and exon 4 of HLF with an insertion of cryptic exon (joining region) maintaining an open-reading frame, whereas type 2 is generated by the fusion between exon 15 of TCF3 and exon 4 of HLF in the same reading frame.[2] |
Characteristic Chromosomal Aberrations / Patterns
Only a few cases have been reported; therefore, no characteristic patterns are noted as of yet. However, ~50% of cases may contain additional abnormalities[10]
Genomic Gain/Loss/LOH
Only a few cases have been reported; therefore, no characteristic patterns are noted as of yet. One case demonstrated loss of 13q12.2q21.1 (containing RB1) and intergenic duplication of NOTCH1.[11] It has described that up to 70% of TCF3::HLF cases have a concurrent PAX5 deletion[3][2]
Gene Mutations (SNV/INDEL)
Very few studies have reported findings, and such studies focus on the specific fusion type. A study comparing t(1;19) (n=29) to t(17;19) (n= 12) cases reported recurrent intragenic deletions of PAX5 or VPREB1 in 2 cases of t(17;19) and RAS signaling pathway genes in 2 cases.[2] Additionally a study of four t(17;19)‐ALL cell lines detected RAS pathway mutations in all four t(17;19)‐ALL lines.[12]
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene (TSG)/Oncogene/Other) | Prevalence (COSMIC/ TCGA/Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance
(Yes, No or Unknown) |
Therapeutic Significance
(Yes, No or Unknown) |
Notes |
| RAS family (NRAS, KRAS, PTPN11, SPHK1); Activating mutations[2] | ||||||||
| TCF3 | One study found TCF3 mutations on the non-fused, normal allele in relapsed/progressing cases.[2] |
Epigenomics (Methylation)
A single study comparing t(17;19) with t(1;19) found 7000 differentially methylated CpG sites between subtypes, of which 78% had lower methylation levels in TCF3::HLF. KBTBD11 differed in methylation and expression between subtypes and before and after remission in TCF3::HLF samples.[13]
Genes and Main Pathways Involved
The chimeric protein from TCF3::HLF acts as a transcription factor resulting in dedifferentiation of mature B lymphocytes.[11] All TCF3 fusions had expression profile signature of B lymphoid cells (PAX5, BLK, CD19, CD22, CD79B, TCF3, EBF1, VPREB1, RAG1, ROR1, BLNK, DNTT). The differences between the TCF3 fusion partners, PBX1, HLF, and ZNF384, led to differential expression patterns based on the genes regulated by these partners. t(17;19) demonstrated induced expression of HLF- regulated genes (SNAI2 (SLUG), GPC4 and BMP3).[2]
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
| TCF3::HLF | Dedifferentiation of mature B lymphocytes[11] |
Diagnostic Testing Methods
- Chromosome analysis
- Fluorescence in situ hybridization (FISH). Break-apart probes may be required to identify cases with cryptic translocations.
- Reverse transcriptase PCR (RT-PCR) may aid in diagnosis.[14]
- Next generation sequencing (NGS)
- Mate pair sequencing (MPseq) to identify the fusion partners in G-band negative, TCF3 BA + cases.[15]
Determination of the fusion partner is necessary as there are significant prognostic differences between the recurrent TCF3::PBX1 fusion and the less common fusions of TCF3::HLF or TCF3::ZNF384.
Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications)
The TCF3::HLF translocation, t (17;19), is associated with a dismal prognosis. A study found t(17;19) may be sensitive to certain drugs in animal models.[2] An in vitro study using four t(17;19)‐ALL and sixteen t(1;19)‐ALL cell lines demonstrated the t(17;19)‐ALL cell lines were more resistant to vincristine (VCR), daunorubicin (DNR), and prednisolone (Pred) than t(1;19)‐ALL cell lines. In addition the t(17;19)‐ALL cell lines were less sensitive to three (Pred, VCR, and l‐asparaginase [l‐Asp]), four (Pred, VCR, l‐Asp, and DNR) and five (Pred, VCR, l‐Asp, DNR, and cyclophosphamide) agents, which are widely used in induction therapy.[12]
An in vitro study demonstrated cells harboring TCF3::HLF are sensitive to PARP inhibitors (olaparib and veliparib)[16], as well as the BCL2 specific inhibitor, venetoclax.[2]
Familial Forms
None reported
Other Information
Put your text here
Links
References
(use "Cite" icon at top of page)
- ↑ 1.0 1.1 1.2 Moorman, Anthony V. (2012-05). "The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia". Blood Reviews. 26 (3): 123–135. doi:10.1016/j.blre.2012.01.001. ISSN 1532-1681. PMID 22436535. Check date values in:
|date=(help) - ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Fischer, Ute; et al. (2015-09). "Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options". Nature Genetics. 47 (9): 1020–1029. doi:10.1038/ng.3362. ISSN 1546-1718. PMC 4603357. PMID 26214592. Check date values in:
|date=(help) - ↑ 3.0 3.1 Akkari, Yassmine M.N.; et al. (2020-05). "Evidence-based review of genomic aberrations in B-lymphoblastic leukemia/lymphoma: Report from the cancer genomics consortium working group for lymphoblastic leukemia". Cancer Genetics. 243: 52–72. doi:10.1016/j.cancergen.2020.03.001. ISSN 2210-7762. Check date values in:
|date=(help) - ↑ 4.0 4.1 4.2 4.3 4.4 Inukai, T.; et al. (2007-02). "Hypercalcemia in childhood acute lymphoblastic leukemia: frequent implication of parathyroid hormone-related peptide and E2A-HLF from translocation 17;19". Leukemia. 21 (2): 288–296. doi:10.1038/sj.leu.2404496. ISSN 0887-6924. PMID 17183364. Check date values in:
|date=(help) - ↑ Borowitz, M. J.; et al. (1993-08-15). "Predictability of the t(1;19)(q23;p13) from surface antigen phenotype: implications for screening cases of childhood acute lymphoblastic leukemia for molecular analysis: a Pediatric Oncology Group study". Blood. 82 (4): 1086–1091. ISSN 0006-4971. PMID 8353275.
- ↑ Borowitz, MJ, et al., (2017). B-lymphoblastic leukaemia/lymphoma with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p129-171.
- ↑ Hiller, B.; et al. (2004-11-16). "CyDAS: a cytogenetic data analysis system". Bioinformatics. 21 (7): 1282–1283. doi:10.1093/bioinformatics/bti146. ISSN 1367-4803.
- ↑ Inaba, T.; et al. (1992-07-24). "Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia". Science (New York, N.Y.). 257 (5069): 531–534. doi:10.1126/science.1386162. ISSN 0036-8075. PMID 1386162.
- ↑ Hunger, S. P. (1996-02-15). "Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: clinical features and molecular pathogenesis". Blood. 87 (4): 1211–1224. ISSN 0006-4971. PMID 8608207.
- ↑ Viguié, F (2011-02). "t(17;19)(q22;p13)". Atlas of Genetics and Cytogenetics in Oncology and Haematology (2). doi:10.4267/2042/37516. ISSN 1768-3262. Check date values in:
|date=(help) - ↑ 11.0 11.1 11.2 Lejman, Monika; et al. (2020-04-03). "Comprehensive chromosomal aberrations in a case of a patient with TCF3-HLF-positive BCP-ALL". BMC medical genomics. 13 (1): 58. doi:10.1186/s12920-020-0709-y. ISSN 1755-8794. PMC 7118981 Check
|pmc=value (help). PMID 32245383 Check|pmid=value (help). - ↑ 12.0 12.1 Watanabe, Atsushi; et al. (2019-07-15). "Resistance of t(17;19)‐acute lymphoblastic leukemia cell lines to multiagents in induction therapy". Cancer Medicine. 8 (11): 5274–5288. doi:10.1002/cam4.2356. ISSN 2045-7634. PMC 6718581. PMID 31305009.
- ↑ Kachroo, Priyadarshini; et al. (2018-02). "NGS-based methylation profiling differentiates TCF3-HLF and TCF3-PBX1 positive B-cell acute lymphoblastic leukemia". Epigenomics. 10 (2): 133–147. doi:10.2217/epi-2017-0080. ISSN 1750-192X. PMID 29334255. Check date values in:
|date=(help) - ↑ Hirabayashi, Shinsuke; et al. (2017-01). "ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype". Haematologica. 102 (1): 118–129. doi:10.3324/haematol.2016.151035. ISSN 1592-8721. PMC 5210242. PMID 27634205. Check date values in:
|date=(help) - ↑ Rowsey, Ross A.; et al. (2019-10-01). "Characterization of TCF3 rearrangements in pediatric B-lymphoblastic leukemia/lymphoma by mate-pair sequencing (MPseq) identifies complex genomic rearrangements and a novel TCF3/TEF gene fusion". Blood Cancer Journal. 9 (10): 81. doi:10.1038/s41408-019-0239-z. ISSN 2044-5385. PMC 6773761. PMID 31575852.
- ↑ Piao, Jinhua; et al. (2017-02-01). "Poly (ADP-ribose) polymerase inhibitors selectively induce cytotoxicity in TCF3-HLF-positive leukemic cells". Cancer Letters. 386: 131–140. doi:10.1016/j.canlet.2016.11.021. ISSN 1872-7980. PMID 27894958.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.