Difference between revisions of "HAEM5:Acute myeloid leukaemia with DEK::NUP214 fusion"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| Line 6: | Line 6: | ||
<blockquote class='blockedit'>{{Box-round|title=HAEM5 Conversion Notes|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Acute Myeloid Leukemia (AML) with t(6;9)(p23;q34.1); DEK-NUP214]]. | <blockquote class='blockedit'>{{Box-round|title=HAEM5 Conversion Notes|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Acute Myeloid Leukemia (AML) with t(6;9)(p23;q34.1); DEK-NUP214]]. | ||
}}</blockquote> | }}</blockquote> | ||
| + | |||
| + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples). Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | ||
| + | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
Revision as of 14:17, 13 December 2023
Haematolymphoid Tumours (5th ed.)
| This page is under construction |
editHAEM5 Conversion NotesThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Acute Myeloid Leukemia (AML) with t(6;9)(p23;q34.1); DEK-NUP214.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples). Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Jennelle C. Hodge, PhD, FACMG
Cancer Category / Type
Cancer Sub-Classification / Subtype
Acute myeloid leukemia (AML) with t(6;9)(p23;q34.1) resulting in DEK-NUP214 fusion
Definition / Description of Disease
This is a distinct entity in the World Health Organization (WHO) classification system, and the most common associated French-American-British (FAB) classifications are M2, M4 and M1[1][2].
Synonyms / Terminology
None
Epidemiology / Prevalence
Accounts for 0.7-1.8% of AML, occurring in both children (median age of 13 years) and adults (median age of 35-44 years)[1].
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table)
| Signs and Symptoms | EXAMPLE Asymptomatic (incidental finding on complete blood counts)
EXAMPLE B-symptoms (weight loss, fever, night sweats) EXAMPLE Fatigue EXAMPLE Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE Cytopenias
EXAMPLE Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.Usually presents with anemia and thrombocytopenia and often with pancytopenia. In adults, the median white blood cell count is 12x10^9/L, which is generally lower than other AML types[1].
Sites of Involvement
Bone marrow
Morphologic Features
This AML subtype may present with or without monocytic features and is frequently associated with basophilia (44-62% of cases) and multilineage dysplasia (most commonly granulocytic and erythroid, and less often megakaryocytic dysplasia)[1]. Auer rods occur in ~1/3 of cases and ringed sideroblasts may also occur[1]. The peripheral blood or bone marrow have >20% blasts characterized by a non-specific myeloid immunophenotype[1].
Immunophenotype
The characteristic immunophenotype of the blasts associated with this entity is listed in the table below. In addition, basophils may present as separate clusters of CD123+, CD33+ CD38+ and HLA-DR- cells[1].
| Finding | Marker |
|---|---|
| Positive (universal) | Myeloperoxidase (MPO), CD9, CD13, CD33, CD38 CD123, and HLA-DR |
| Positive (subset) | KIT(CD117), CD34, CD15, CD64 (monocyte-associated marker), and Dexoynucleotidyl Transferase (TdT) |
| Negative (universal) | |
| Negative (subset) |
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE t(9;22)(q34;q11.2) | EXAMPLE 3'ABL1 / 5'BCR | EXAMPLE der(22) | EXAMPLE 20% (COSMIC)
EXAMPLE 30% (add reference) |
Yes | No | Yes | EXAMPLE
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
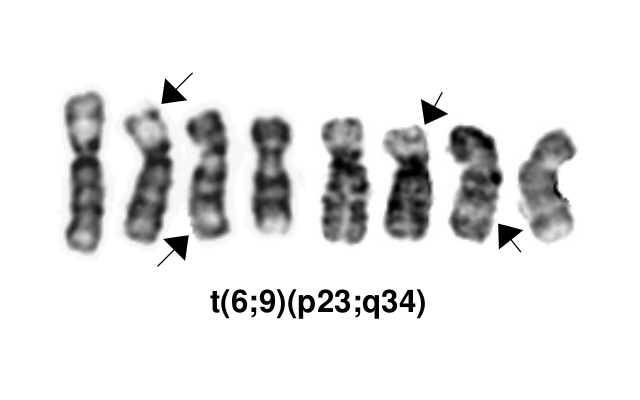
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.This AML subtype is classified based on the presence of a t(6;9)(p23;q34.1), which results in fusion of the 5’ portion of DEK at “6p23” (specifically 6p22.3[hg38]) and the 3’ portion of NUP214(CAN) at “9q34.1” (specifically 9q34.13[hg38]). The breakpoints are intronic, producing an in-frame fusion[3]. The DEK-NUP214 fusion present on the derivative chromosome 6 is considered the pathogenic entity as the reciprocal NUP214-DEK fusion on chromosome 9 does not appear to be transcribed[4]. Typically the DEK-NUP214 fusion presents as the sole abnormality but can be part of a complex karyotype[1].
Chromosomal Rearrangement Genes in Fusion (5’ or 3’ Segments) Pathogenic Derivative Prevalence t(6;9)(p23;q34.1) 5'DEK / 3'NUP214(CAN) der(6) 0.7-1.8% of AML
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
This translocation has traditionally been associated with a poor prognosis in both adult and pediatric cases[1]. Of note, a 2014 retrospective analysis suggests a better outcome for pediatric patients with this translocation than previously reported[5]. Elevated white blood cell counts and higher bone marrow blast percentages are associated with shorter periods of overall survival and disease-free survival, respectively[1].
Limited data suggests early allogeneic stem cell transplantation may be associated with better overall survival compared to patients without transplantation, suggesting accurate diagnosis for these patients is crucial[1][6][7].
The concurrent presence of FLT3-ITD does not appear to negatively impact survival in the pediatric population[1].
Cases with the 6;9 translocation and <20% blasts are not currently classified as AML, which is controversial. Such cases should have close follow-up to monitor for development of more definitive evidence of AML or may be treated as AML if clinically appropriate[1].
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE
7 |
EXAMPLE Loss | EXAMPLE
chr7:1- 159,335,973 [hg38] |
EXAMPLE
chr7 |
Yes | Yes | No | EXAMPLE
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE
8 |
EXAMPLE Gain | EXAMPLE
chr8:1-145,138,636 [hg38] |
EXAMPLE
chr8 |
No | No | No | EXAMPLE
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.Not applicable
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.Not applicable
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.COSMIC does not have specific information on mutations related to this subtype of AML.
Other Mutations
Type Gene/Region/Other Concomitant Mutations FLT3-ITD (69% of children and 78% of adults) Secondary Mutations Mutually Exclusive FLT3-TKD is very uncommon
Epigenomic Alterations
Not applicable
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.The molecular mechanism is not completely understood, but the fusion protein is known to act as an aberrant transcription factor, alter nuclear transport and induce myeloid cell-specific global protein synthesis[1][8].
Genetic Diagnostic Testing Methods
Karyotype, FISH, RT-PCR
Familial Forms
Not applicable
Additional Information
Not applicable
Links
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p137-138.
- ↑ Oyarzo, Mauricio P.; et al. (2004). "Acute myeloid leukemia with t(6;9)(p23;q34) is associated with dysplasia and a high frequency of flt3 gene mutations". American Journal of Clinical Pathology. 122 (3): 348–358. doi:10.1309/5DGB-59KQ-A527-PD47. ISSN 0002-9173. PMID 15362364.
- ↑ von Lindern, M.; et al. (1992). "The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA". Molecular and Cellular Biology. 12 (4): 1687–1697. doi:10.1128/mcb.12.4.1687. ISSN 0270-7306. PMC 369612. PMID 1549122.CS1 maint: PMC format (link)
- ↑ von Lindern, M.; et al. (1992). "Translocation t(6;9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene". Bailliere's Clinical Haematology. 5 (4): 857–879. doi:10.1016/s0950-3536(11)80049-1. ISSN 0950-3536. PMID 1308167.
- ↑ Sandahl, Julie Damgaard; et al. (2014). "t(6;9)(p22;q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients". Haematologica. 99 (5): 865–872. doi:10.3324/haematol.2013.098517. ISSN 1592-8721. PMC 4008104. PMID 24441146.
- ↑ Slovak, M. L.; et al. (2006). "A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare 'poor prognosis' myeloid malignancies". Leukemia. 20 (7): 1295–1297. doi:10.1038/sj.leu.2404233. ISSN 0887-6924. PMID 16628187.
- ↑ Ishiyama, K.; et al. (2012). "Allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia with t(6;9)(p23;q34) dramatically improves the patient prognosis: a matched-pair analysis". Leukemia. 26 (3): 461–464. doi:10.1038/leu.2011.229. ISSN 1476-5551. PMID 21869835.
- ↑ Ageberg, Malin; et al. (2008). "Identification of a novel and myeloid specific role of the leukemia-associated fusion protein DEK-NUP214 leading to increased protein synthesis". Genes, Chromosomes & Cancer. 47 (4): 276–287. doi:10.1002/gcc.20531. ISSN 1098-2264. PMID 18181180.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Acute myeloid leukaemia with DEK::NUP214 fusion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 12/13/2023, https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_DEK::NUP214_fusion.