Difference between revisions of "ABL1"
| [unchecked revision] | [unchecked revision] |
| Line 1: | Line 1: | ||
| − | + | ==Primary Author(s)*== | |
| + | |||
| + | Put your text here | ||
| + | |||
| + | __TOC__ | ||
| + | |||
| + | ==Synonyms== | ||
| + | |||
| + | Put your text here | ||
| + | |||
| + | EXAMPLE: Tumor protein p53, ''LFS1, p53, BCC7, TRP53'' | ||
| + | |||
| + | ==Genomic Location== | ||
| + | |||
| + | '''Cytoband:''' Put your text here. EXAMPLE: 17p13.1 | ||
| + | |||
| + | '''Genomic Coordinates:''' | ||
| + | |||
| + | Put your text here | ||
| + | |||
| + | EXAMPLE: chr17:7,571,720-7,590,868 [hg19] | ||
| + | |||
| + | EXAMPLE: chr17:7,668,402-7,687,538 [hg38] | ||
| + | |||
| + | ==Cancer Category/Type== | ||
| + | |||
| + | Put your text here | ||
| + | |||
| + | ==Gene Overview== | ||
| + | |||
| + | Put your text here. | ||
| + | |||
| + | ==Common Alteration Types== | ||
| + | |||
| + | Put your text here and/or fill in the table with an X where applicable | ||
| + | |||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | ! Copy Number Loss !! Copy Number Gain !! LOH !! Loss-of-Function Mutation !! Gain-of-Function Mutation !! Translocation/Fusion | ||
| + | |- | ||
| + | | EXAMPLE: X ||EXAMPLE: X || EXAMPLE: X || EXAMPLE: X || EXAMPLE: X || EXAMPLE: X | ||
| + | |} | ||
| + | |||
| + | ==Internal Pages== | ||
| + | |||
| + | Put your text here | ||
| + | |||
| + | EXAMPLE [[Germline Cancer Predisposition Genes]] | ||
| + | |||
| + | ==External Links== | ||
| + | |||
| + | Put your text here - Include as applicable links to: 1) Atlas of Genetics and Cytogenetics in Oncology and Haematology, 2) COSMIC, 3) CIViC, 4) St. Jude ProteinPaint, 5) Precision Medicine Knnowledgebase (Weill Cornell), 6) Cancer Index, 7) OncoKB, 8) My Cancer Genome, 9) UniProt, 10) Pfam, 11) GeneCards, 12) GeneReviews, and 13) Any gene-specific databases. | ||
| + | |||
| + | EXAMPLES | ||
| + | |||
| + | '''[http://atlasgeneticsoncology.org/Genes/P53ID88.html ''TP53'' by Atlas of Genetics and Cytogenetics in Oncology and Haematology]''' - detailed gene information | ||
| + | |||
| + | '''[https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=TP53 ''TP53'' by COSMIC]''' - sequence information, expression, catalogue of mutations | ||
| + | |||
| + | '''[https://civicdb.org/events/genes/45/summary/variants/1300/summary ''TP53'' by CIViC]''' - general knowledge and evidence-based variant specific information | ||
| + | |||
| + | '''[http://p53.iarc.fr/ ''TP53'' by IARC]''' - ''TP53'' database with reference sequences and mutational landscape | ||
| + | |||
| + | '''[https://pecan.stjude.cloud/proteinpaint/tp53 ''TP53'' by St. Jude ProteinPaint]''' mutational landscape and matched expression data. | ||
| + | |||
| + | '''[https://pmkb.weill.cornell.edu/search?utf8=%E2%9C%93&search=tp53 ''TP53'' by Precision Medicine Knowledgebase (Weill Cornell)]''' - manually vetted interpretations of variants and CNVs | ||
| + | |||
| + | '''[http://www.cancerindex.org/geneweb/TP53.htm ''TP53'' by Cancer Index]''' - gene, pathway, publication information matched to cancer type | ||
| + | |||
| + | '''[http://oncokb.org/#/gene/TP53 ''TP53'' by OncoKB]''' - mutational landscape, mutation effect, variant classification | ||
| + | |||
| + | '''[https://www.mycancergenome.org/content/gene/tp53/ ''TP53'' by My Cancer Genome]''' - brief gene overview | ||
| + | |||
| + | '''[http://www.uniprot.org/uniprot/P04637 ''TP53'' by UniProt]''' - protein and molecular structure and function | ||
| + | |||
| + | '''[https://pfam.xfam.org/family/p53 ''TP53'' by Pfam]''' - gene and protein structure and function information | ||
| + | |||
| + | '''[http://www.genecards.org/cgi-bin/carddisp.pl?gene=tp53 ''TP53'' by GeneCards]''' - general gene information and summaries | ||
| + | |||
| + | '''[https://www.ncbi.nlm.nih.gov/books/NBK1311/ GeneReviews]''' - information on Li Fraumeni Syndrome | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | === EXAMPLE Book === | ||
| + | #Arber DA, et al., (2008). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, Editors. IARC Press: Lyon, France, p117-118. | ||
| + | |||
| + | === EXAMPLE Journal Article === | ||
| + | #Li Y, et al., (2001). Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 28:220-221, PMID 11431691. | ||
| + | |||
| + | == Notes == | ||
| + | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. | ||
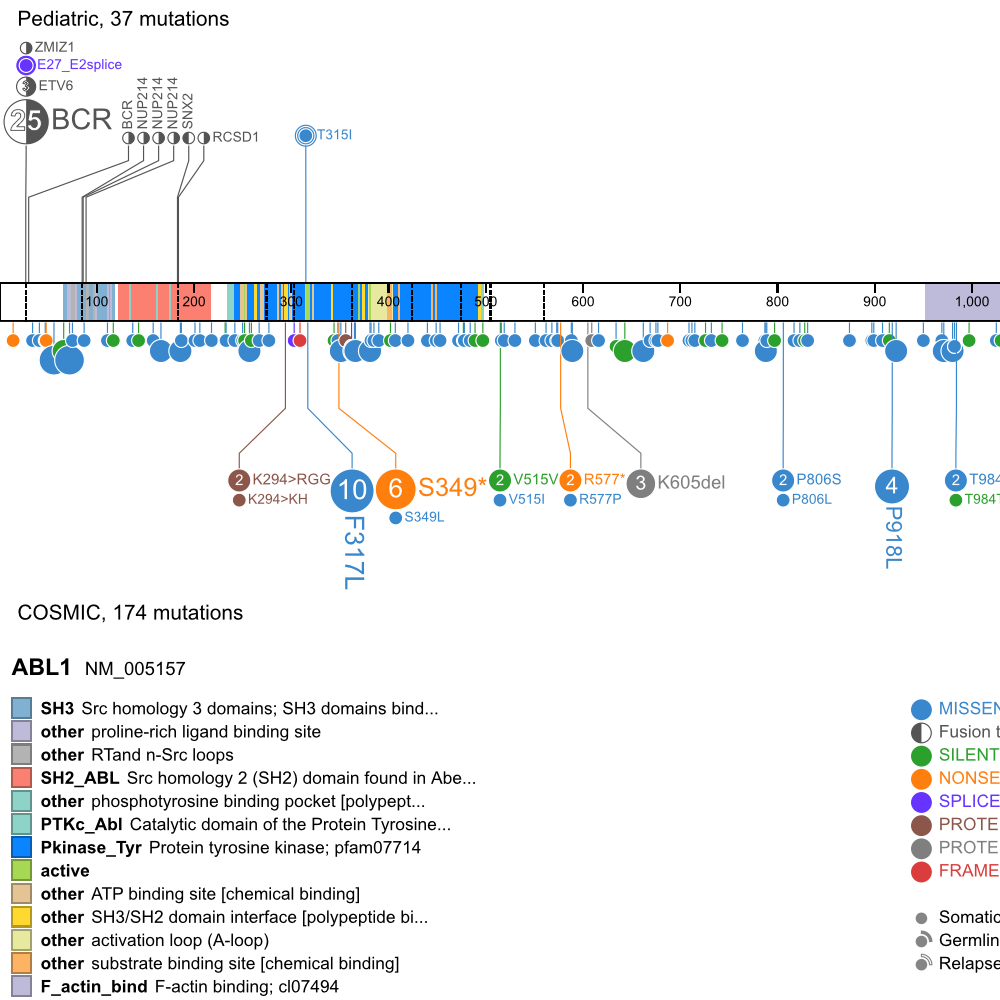
[[File:ABL1.png|frame|50px|link=https://pecan.stjude.org/proteinpaint/ABL1]] | [[File:ABL1.png|frame|50px|link=https://pecan.stjude.org/proteinpaint/ABL1]] | ||
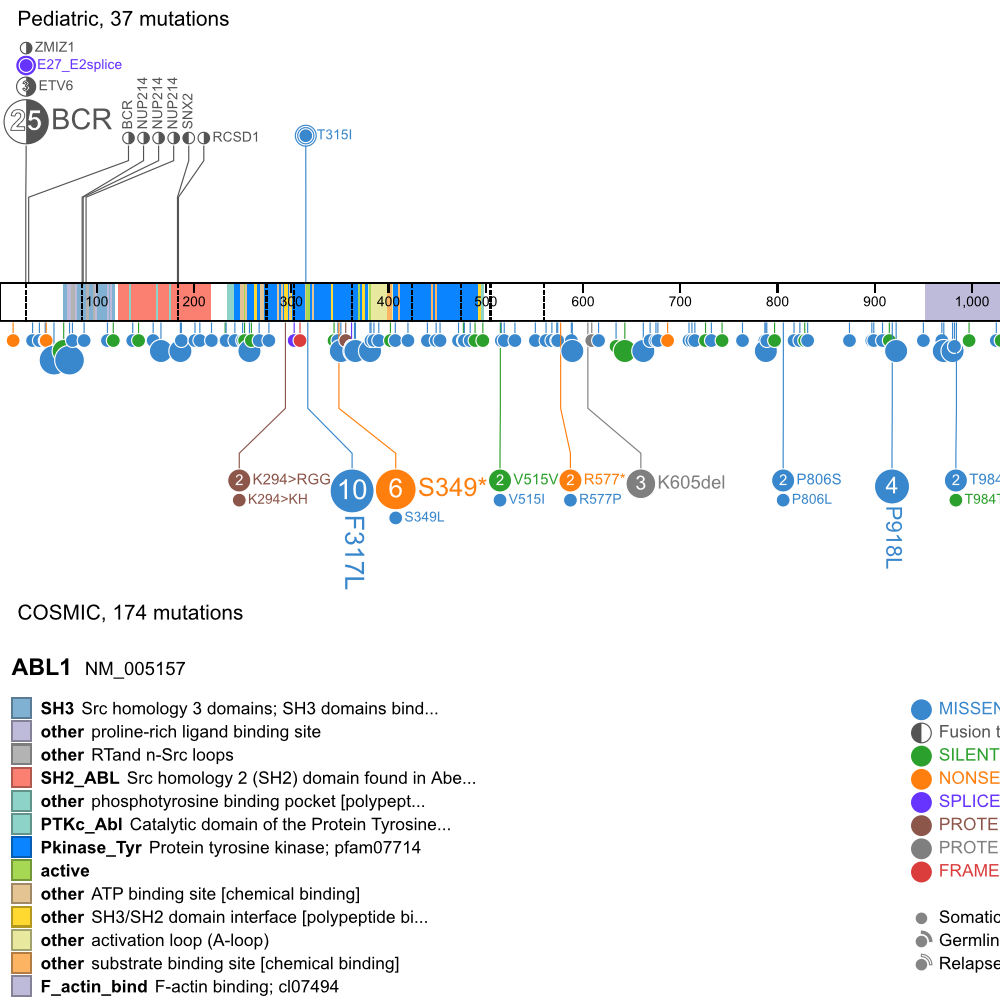
[[File:ABL1.V2.png|frame|50px|link=https://pecan.stjude.org/proteinpaint/ABL1]] | [[File:ABL1.V2.png|frame|50px|link=https://pecan.stjude.org/proteinpaint/ABL1]] | ||
Revision as of 22:14, 16 May 2018
Primary Author(s)*
Put your text here
Synonyms
Put your text here
EXAMPLE: Tumor protein p53, LFS1, p53, BCC7, TRP53
Genomic Location
Cytoband: Put your text here. EXAMPLE: 17p13.1
Genomic Coordinates:
Put your text here
EXAMPLE: chr17:7,571,720-7,590,868 [hg19]
EXAMPLE: chr17:7,668,402-7,687,538 [hg38]
Cancer Category/Type
Put your text here
Gene Overview
Put your text here.
Common Alteration Types
Put your text here and/or fill in the table with an X where applicable
| Copy Number Loss | Copy Number Gain | LOH | Loss-of-Function Mutation | Gain-of-Function Mutation | Translocation/Fusion |
|---|---|---|---|---|---|
| EXAMPLE: X | EXAMPLE: X | EXAMPLE: X | EXAMPLE: X | EXAMPLE: X | EXAMPLE: X |
Internal Pages
Put your text here
EXAMPLE Germline Cancer Predisposition Genes
External Links
Put your text here - Include as applicable links to: 1) Atlas of Genetics and Cytogenetics in Oncology and Haematology, 2) COSMIC, 3) CIViC, 4) St. Jude ProteinPaint, 5) Precision Medicine Knnowledgebase (Weill Cornell), 6) Cancer Index, 7) OncoKB, 8) My Cancer Genome, 9) UniProt, 10) Pfam, 11) GeneCards, 12) GeneReviews, and 13) Any gene-specific databases.
EXAMPLES
TP53 by Atlas of Genetics and Cytogenetics in Oncology and Haematology - detailed gene information
TP53 by COSMIC - sequence information, expression, catalogue of mutations
TP53 by CIViC - general knowledge and evidence-based variant specific information
TP53 by IARC - TP53 database with reference sequences and mutational landscape
TP53 by St. Jude ProteinPaint mutational landscape and matched expression data.
TP53 by Precision Medicine Knowledgebase (Weill Cornell) - manually vetted interpretations of variants and CNVs
TP53 by Cancer Index - gene, pathway, publication information matched to cancer type
TP53 by OncoKB - mutational landscape, mutation effect, variant classification
TP53 by My Cancer Genome - brief gene overview
TP53 by UniProt - protein and molecular structure and function
TP53 by Pfam - gene and protein structure and function information
TP53 by GeneCards - general gene information and summaries
GeneReviews - information on Li Fraumeni Syndrome
References
EXAMPLE Book
- Arber DA, et al., (2008). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, Editors. IARC Press: Lyon, France, p117-118.
EXAMPLE Journal Article
- Li Y, et al., (2001). Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 28:220-221, PMID 11431691.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.