Difference between revisions of "HAEM5:Acute myeloid leukaemia with NUP98 rearrangement"
| [unchecked revision] | [pending revision] |
(Added the text details) |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
{{Under Construction}} | {{Under Construction}} | ||
| − | + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | |
| − | |||
| − | |||
| − | |||
| − | ( | ||
| − | + | ==Primary Author(s)*== | |
Eric McGinnis, MD | Eric McGinnis, MD | ||
| Line 17: | Line 13: | ||
Fatma Albulushi, MD | Fatma Albulushi, MD | ||
| + | __TOC__ | ||
| − | + | ==WHO Classification of Disease== | |
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
| + | !Structure | ||
| + | !Disease | ||
| + | |- | ||
|Book | |Book | ||
|Haematolymphoid Tumours (5th ed.) | |Haematolymphoid Tumours (5th ed.) | ||
|- | |- | ||
|Category | |Category | ||
| − | |Myeloid | + | |Myeloid proliferations and neoplasms |
|- | |- | ||
|Family | |Family | ||
| − | |Acute | + | |Acute myeloid leukaemia |
|- | |- | ||
|Type | |Type | ||
| − | |Acute | + | |Acute myeloid leukaemia with defining genetic abnormalities |
|- | |- | ||
|Subtype(s) | |Subtype(s) | ||
| − | |Acute | + | |Acute myeloid leukaemia with NUP98 rearrangement |
|} | |} | ||
| − | + | ==WHO Essential and Desirable Genetic Diagnostic Criteria.== | |
| − | |||
| − | |||
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
|WHO Essential Criteria (Genetics)* | |WHO Essential Criteria (Genetics)* | ||
| Line 54: | Line 50: | ||
<nowiki>*</nowiki>Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the WHO Classification of Tumours. | <nowiki>*</nowiki>Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the WHO Classification of Tumours. | ||
| + | ==Definition / Description of Disease== | ||
| + | |||
| + | Put your text here <span style="color:#0070C0">(''Instructions: Brief description of approximately one paragraph - include disease context relative to other WHO classification categories, diagnostic criteria if applicable, and differential diagnosis if applicable. Other classifications can be referenced for comparison.'') </span> | ||
| + | |||
| + | ==Synonyms / Terminology== | ||
| + | |||
| + | Put your text here <span style="color:#0070C0">(''Instructions: Include currently used terms and major historical ones, adding “(historical)” after the latter.'') </span> | ||
| + | |||
| + | ==Epidemiology / Prevalence== | ||
| + | |||
| + | Put your text here | ||
| − | + | ==Clinical Features== | |
| − | ('' | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
| − | | | + | |'''Signs and Symptoms''' |
| − | | | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| + | |||
| + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) | ||
| + | |||
| + | <span class="blue-text">EXAMPLE:</span> Fatigue | ||
| + | |||
| + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) | ||
|- | |- | ||
| − | | | + | |'''Laboratory Findings''' |
| − | | | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| + | |||
| + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) | ||
|} | |} | ||
| + | ==Sites of Involvement== | ||
| + | |||
| + | Put your text here <span style="color:#0070C0">(''Instruction: Indicate physical sites; <span class="blue-text">EXAMPLE:</span> nodal, extranodal, bone marrow'') </span> | ||
| − | + | ==Morphologic Features== | |
| − | + | Put your text here | |
| + | ==Immunophenotype== | ||
| − | The NUP98 gene (chromosome 11p15) encodes a nucleoporin protein, which is part of the nuclear pore complex which regulates nucleocytoplasmic transport of protein and RNA. NUP98 fusion proteins involve the N-terminal portion of NUP98 and the C-terminal portion of the fusion partner. These fusion partners consist of homeodomain proteins, which are transcription factors, and non-homeodomain proteins, which are thought to play a role in transcriptional or epigenetic regulation. <ref name=":0" /><ref name=":1">{{Cite journal|last=Bertrums|first=Eline J. M.|last2=Smith|first2=Jenny L.|last3=Harmon|first3=Lauren|last4=Ries|first4=Rhonda E.|last5=Wang|first5=Yi-Cheng J.|last6=Alonzo|first6=Todd A.|last7=Menssen|first7=Andrew J.|last8=Chisholm|first8=Karen M.|last9=Leonti|first9=Amanda R.|date=2023-02-23|title=Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia|url=https://www.haematologica.org/article/view/haematol.2022.281653|journal=Haematologica|language=en|volume=108|issue=8|pages=2044–2058|doi=10.3324/haematol.2022.281653|issn=1592-8721}}</ref> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
| + | |||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Finding!!Marker | ||
| + | |- | ||
| + | |Positive (universal)||<span class="blue-text">EXAMPLE:</span> CD1 | ||
| + | |- | ||
| + | |Positive (subset)||<span class="blue-text">EXAMPLE:</span> CD2 | ||
| + | |- | ||
| + | |Negative (universal)||<span class="blue-text">EXAMPLE:</span> CD3 | ||
| + | |- | ||
| + | |Negative (subset)||<span class="blue-text">EXAMPLE:</span> CD4 | ||
| + | |} | ||
| + | |||
| + | ==Chromosomal Rearrangements (Gene Fusions)== | ||
| + | |||
| + | Acute myeloid leukaemia (AML) with NUP98 rearrangement is characterized by chromosomal translocations involving NUP98 (nucleoporin 98 kDa) on chromosome 11p15.4 and various partner genes. (Reference WHO book). There are over 40 fusion partners which have been reported to date. NUP98 fusions can be categorized into three broad parts. The first category includes NUP98 fusions with transcription factors as partners, which can change the expression of target genes through DNA binding domains. The second category is NUP98 fusions with epigenetic modifiers that modify chromatin to change target gene expression. The third category of NUP98 fusions has neither the DNA binding nor chromatin remodeling domain.<ref name=":0">{{Cite journal|last=Mohanty|first=Sagarajit|date=2023-09|title=NUP98 Rearrangements in AML: Molecular Mechanisms and Clinical Implications|url=https://www.mdpi.com/2673-7523/3/3/11|journal=Onco|language=en|volume=3|issue=3|pages=147–164|doi=10.3390/onco3030011|issn=2673-7523}}</ref> | ||
| + | |||
| + | |||
| + | The NUP98 gene (chromosome 11p15) encodes a nucleoporin protein, which is part of the nuclear pore complex which regulates nucleocytoplasmic transport of protein and RNA. NUP98 fusion proteins involve the N-terminal portion of NUP98 and the C-terminal portion of the fusion partner. These fusion partners consist of homeodomain proteins, which are transcription factors, and non-homeodomain proteins, which are thought to play a role in transcriptional or epigenetic regulation.<ref name=":0" /><ref name=":1">{{Cite journal|last=Bertrums|first=Eline J. M.|last2=Smith|first2=Jenny L.|last3=Harmon|first3=Lauren|last4=Ries|first4=Rhonda E.|last5=Wang|first5=Yi-Cheng J.|last6=Alonzo|first6=Todd A.|last7=Menssen|first7=Andrew J.|last8=Chisholm|first8=Karen M.|last9=Leonti|first9=Amanda R.|date=2023-02-23|title=Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia|url=https://www.haematologica.org/article/view/haematol.2022.281653|journal=Haematologica|language=en|volume=108|issue=8|pages=2044–2058|doi=10.3324/haematol.2022.281653|issn=1592-8721}}</ref> | ||
<br /> | <br /> | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 80: | Line 119: | ||
|'''Typical Chromosomal Alteration(s)''' | |'''Typical Chromosomal Alteration(s)''' | ||
|'''Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' | |'''Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' | ||
| − | |'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' | + | |'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' |
|'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | |'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
|'''Clinical Relevance Details/Other Notes''' | |'''Clinical Relevance Details/Other Notes''' | ||
| Line 87: | Line 126: | ||
|''NUP98::NSD1'' | |''NUP98::NSD1'' | ||
<br /> | <br /> | ||
| − | |NUP98-NSD1 prevents EZH2-mediated repression of Hox-A locus genes by colocalizing H3K36 methylation and histone acetylation at regulatory DNA elements hence preventing myeloid progenitor | + | |NUP98-NSD1 prevents EZH2-mediated repression of Hox-A locus genes by colocalizing H3K36 methylation and histone acetylation at regulatory DNA elements hence preventing myeloid progenitor immortalization. |
|t(5;11)(q35;p15) | |t(5;11)(q35;p15) | ||
| Line 94: | Line 133: | ||
|Defining genetic abnormality in AML | |Defining genetic abnormality in AML | ||
|Yes (WHO) | |Yes (WHO) | ||
| − | |Rare but recurrent alteration seen mainly in children and young adults with AML. Poor overall survival, disease free survival, induction failure and chemotherapy resistance <ref name=":1" /> | + | |Rare but recurrent alteration seen mainly in children and young adults with AML. Poor overall survival, disease free survival, induction failure and chemotherapy resistance.<ref name=":1" /> |
|- | |- | ||
|''NUP98'' | |''NUP98'' | ||
|''NUP98::KDM5A'' | |''NUP98::KDM5A'' | ||
| − | |KDM5A is an epigenetic-modifying partners of NUP98 which dysregulate Hox genes expression through recognition of H3K4me3/2 marks by the plant homeodomain (PHD) finger domain | + | |KDM5A is an epigenetic-modifying partners of NUP98 which dysregulate Hox genes expression through recognition of H3K4me3/2 marks by the plant homeodomain (PHD) finger domain. |
|t(11;12)(p15;p13) | |t(11;12)(p15;p13) | ||
| Line 105: | Line 144: | ||
|Defining genetic abnormality in AML | |Defining genetic abnormality in AML | ||
|Yes (WHO) | |Yes (WHO) | ||
| − | |''Commonly associated with erythroid and megakaryocytic phenotypes in | + | |''Commonly associated with erythroid and megakaryocytic phenotypes in pediatric AML (acute erythroid leukemia and acute megakaryocytic leukemia).''<ref name=":1" /> |
| − | '' | + | ''Usually associate with unfavorable outcomes'' |
|- | |- | ||
|''NUP98'' | |''NUP98'' | ||
|''NUP98::HOXA9'' | |''NUP98::HOXA9'' | ||
<br /> | <br /> | ||
| − | |NUP98 fusions bind near the HOX genes loci and activate their expression through chromatin remodeling. The overexpression of distal HoxA cluster genes promote self-renewal and drive leukogenesis | + | |NUP98 fusions bind near the HOX genes loci and activate their expression through chromatin remodeling. The overexpression of distal HoxA cluster genes promote self-renewal and drive leukogenesis. |
|t(7;11)(p15, p15) | |t(7;11)(p15, p15) | ||
|Rare (AML) | |Rare (AML) | ||
| Line 119: | Line 158: | ||
| | | | ||
|} | |} | ||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Chromosomal Rearrangement!!Genes in Fusion (5’ or 3’ Segments)!!Pathogenic Derivative!!Prevalence | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | ||
| + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) | ||
| + | |Yes | ||
| + | |No | ||
| + | |Yes | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | |
| − | + | |} | |
| + | |||
| + | ==Individual Region Genomic Gain / Loss / LOH== | ||
No characteristic chromosomal gain or loss. However, trisomy 8 and chromosome 13 abnormalities may be observed. | No characteristic chromosomal gain or loss. However, trisomy 8 and chromosome 13 abnormalities may be observed. | ||
| − | Several reports indicated that del(13q) is a frequent event in ''NUP98::KDM5A'' AML patients, indicating co-occurrence of ''NUP98-KDMA'' fusion with ''RB1'' deletion. | + | Several reports indicated that del(13q) is a frequent event in ''NUP98::KDM5A'' AML patients, indicating co-occurrence of ''NUP98-KDMA'' fusion with ''RB1'' deletion. |
<br /> | <br /> | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | |'''Chromosome Number''' | + | |'''Chromosome Number''' |
|'''Gain/Loss/Amp/LOH''' | |'''Gain/Loss/Amp/LOH''' | ||
|'''Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size]''' | |'''Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size]''' | ||
|'''Relevant Gene(s)''' | |'''Relevant Gene(s)''' | ||
| − | |'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' | + | |'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' |
|'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | |'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
|'''Clinical Relevance Details/Other Notes''' | |'''Clinical Relevance Details/Other Notes''' | ||
| Line 140: | Line 195: | ||
|Trisomy 8 | |Trisomy 8 | ||
|Unknown | |Unknown | ||
| − | |NA | + | |NA |
|No | |No | ||
| | | | ||
| Line 151: | Line 206: | ||
| | | | ||
|Particularly associated with NUP98::KDM5A | |Particularly associated with NUP98::KDM5A | ||
| − | |} | + | |} |
| + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> | ||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Chr #!!Gain / Loss / Amp / LOH!!Minimal Region Genomic Coordinates [Genome Build]!!Minimal Region Cytoband | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | |||
| + | 7 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Loss | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | chr7:1- 159,335,973 [hg38] | |
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | chr7 | |
| + | |Yes | ||
| + | |Yes | ||
| + | |No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
| − | + | 8 | |
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
| − | |EXAMPLE: | + | |
| − | | | + | chr8:1-145,138,636 [hg38] |
| − | | | + | |<span class="blue-text">EXAMPLE:</span> |
| − | | | + | |
| − | |EXAMPLE: | + | chr8 |
| + | |No | ||
| + | |No | ||
| + | |No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Common recurrent secondary finding for t(8;21) (add reference). | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| + | ==Characteristic Chromosomal Patterns== | ||
| + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> | ||
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Chromosomal Pattern | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Co-deletion of 1p and 18q | |
| + | |Yes | ||
| + | |No | ||
| + | |No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | Loss of RB1 at 13q14 is particularly associated with NUP98::KDM5A | + | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
| + | |} | ||
| + | ==Gene Mutations (SNV / INDEL)== | ||
| + | FLT3-ITD and WT1 mutation are recurring events in NUP98::NSD1 and was also observed in some NUP98::HOXA9 AML patients.(R1). Loss of RB1 at 13q14 is particularly associated with NUP98::KDM5A | ||
{| class="wikitable" | {| class="wikitable" | ||
|'''Gene''' | |'''Gene''' | ||
| Line 202: | Line 279: | ||
|'''Tumor Suppressor Gene (TSG)/Oncogene/Other''' | |'''Tumor Suppressor Gene (TSG)/Oncogene/Other''' | ||
|'''Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' | |'''Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' | ||
| − | |'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' | + | |'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' |
|'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | |'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
|'''Clinical Relevance Details/Other Notes''' | |'''Clinical Relevance Details/Other Notes''' | ||
| Line 209: | Line 286: | ||
| | | | ||
| | | | ||
| + | |Recurrent | ||
| + | |Poor prognosis | ||
| | | | ||
| − | | | + | |Seen in 67 to 91% of cases with NUP98::NSD1 |
| − | |||
| − | |||
|- | |- | ||
|''WT1'' | |''WT1'' | ||
| | | | ||
| | | | ||
| + | |Rare | ||
| | | | ||
| | | | ||
| − | | | + | |Reported in 33-55% of NUP98::NSD1 rearranged AML |
| − | |||
|- | |- | ||
|''RB1'' | |''RB1'' | ||
| | | | ||
| | | | ||
| + | |Rare | ||
| | | | ||
| | | | ||
| − | | | + | |Particularly associated with NUP98::KDM5A |
| − | |||
|} | |} | ||
| − | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> | |
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Gene; Genetic Alteration!!'''Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other)'''!!'''Prevalence (COSMIC / TCGA / Other)'''!!'''Concomitant Mutations'''!!'''Mutually Exclusive Mutations''' | ||
| + | !'''Diagnostic Significance (Yes, No or Unknown)''' | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations | ||
| − | + | <span class="blue-text">EXAMPLE:</span> | |
| − | + | EGFR; Exon 20 mutations | |
| − | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations | |
| + | |<span class="blue-text">EXAMPLE:</span> TSG | ||
| + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | ||
| − | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) | |
| − | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H | |
| − | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | | | ||
| | | | ||
| | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). | ||
| + | <br /> | ||
|} | |} | ||
| + | Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | ||
| + | ==Epigenomic Alterations== | ||
| − | + | Put your text here | |
| − | + | ==Genes and Main Pathways Involved== | |
| − | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> | |
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling | ||
| + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation | ||
| + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling | ||
| + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program | ||
| + | |} | ||
| + | ==Genetic Diagnostic Testing Methods== | ||
| − | + | Rearrangements involving NUP98 are often cryptic on conventional karyotype, owing to terminal location of NUP98 on chromosome 11p15.4. Most patients have a normal karyotype. Diagnosis is established using the following tests: | |
| − | + | * FISH using NUP98 break-apart probes | |
| + | * RT-PCR for fusion proteins like NUP98::NSD1 | ||
| + | * RNA sequencing | ||
| + | * Optical Genome Mapping (OGM) | ||
| − | + | <br /> | |
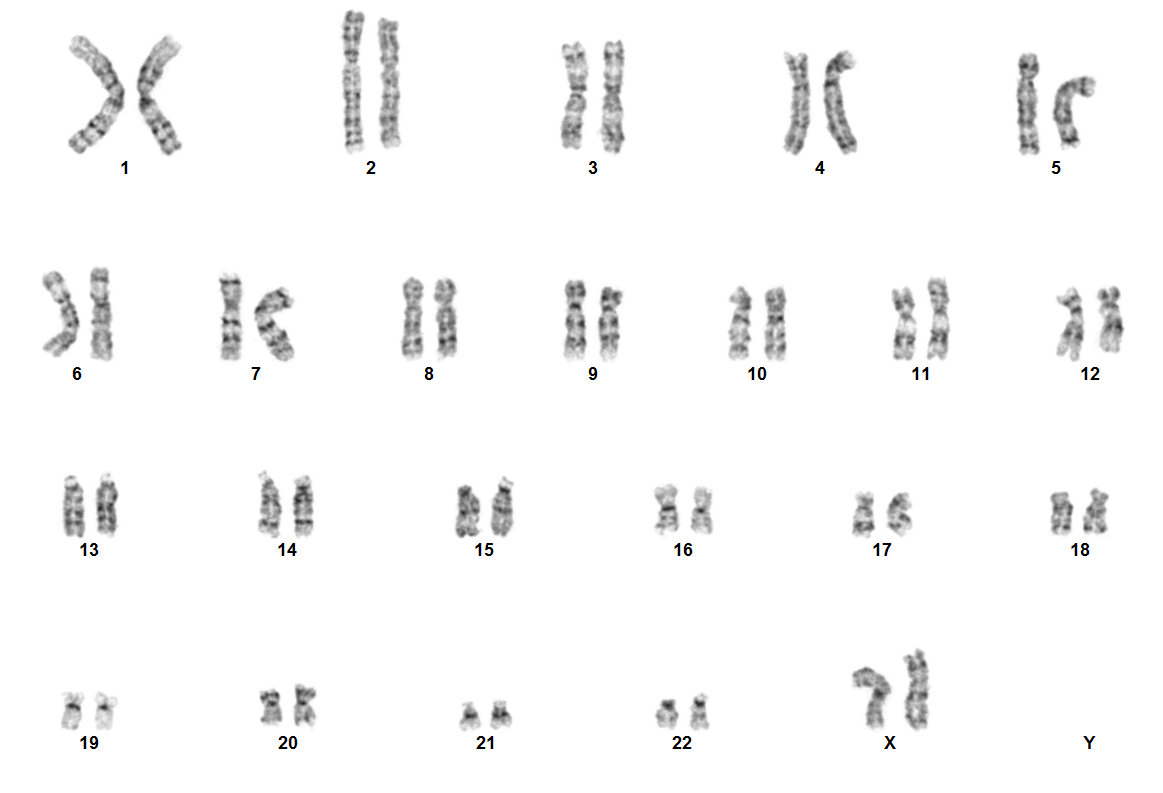
| + | [[File:NUP98 NSD1.png|none|thumb|617x617px|Karyotype image of NUP98 rearranged acute myeloid leukemia. Due to the cryptic nature of NUP98 rearrangement, karyotype is usually normal. ]] | ||
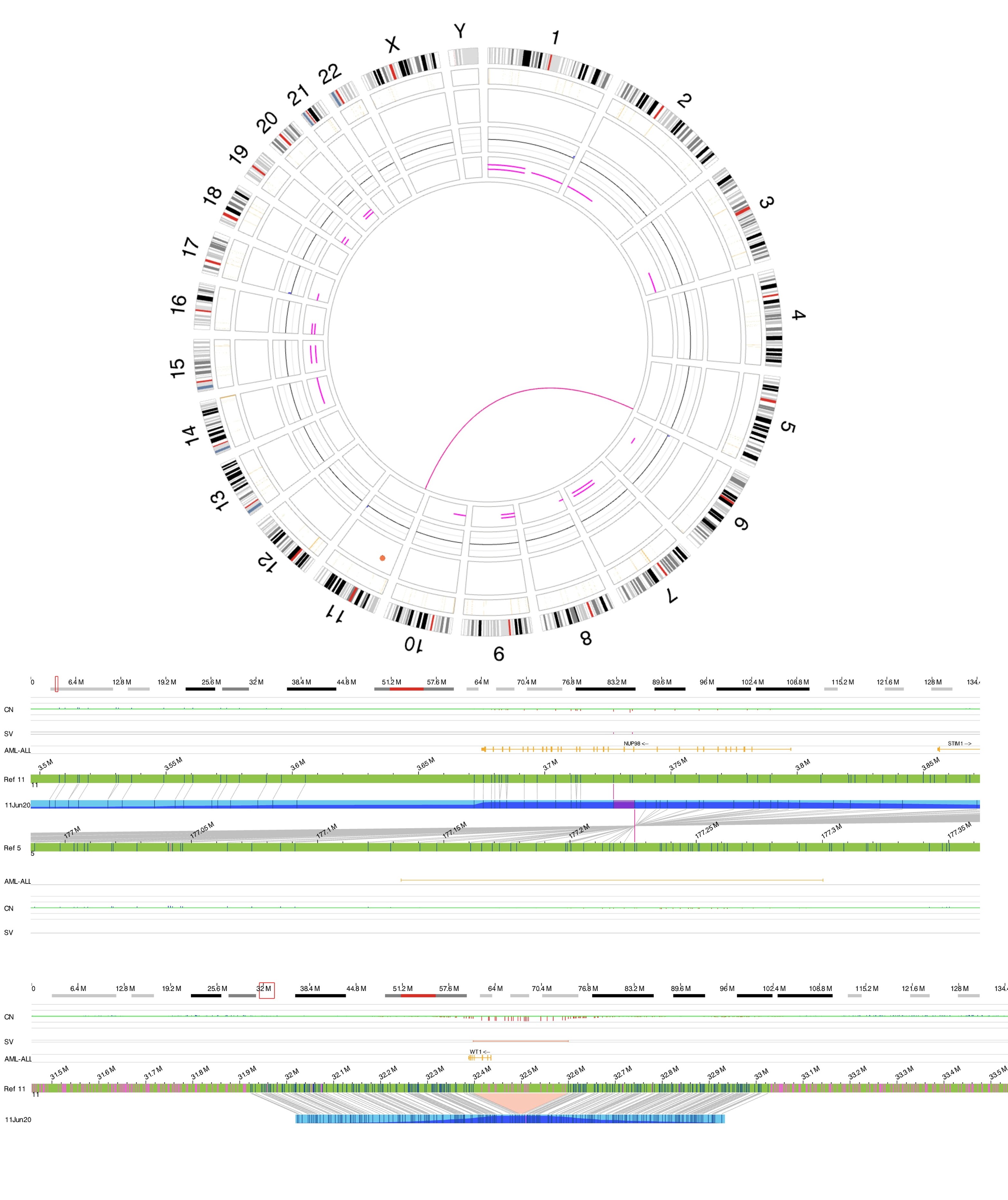
| + | [[File:T(5;11).jpg|none|thumb|584x584px|Optical genome mapping. Figure A showing circus plot with t(5;11). Figure B showing exact breakpoints of the translocation leading to NUP98::NSD1 fusion. Figure C showing WT1 deletion which is a common secondary event in NUP98 rearranged AML.]] | ||
| + | <br /> | ||
| − | + | ==Familial Forms== | |
| + | Put your text here <span style="color:#0070C0">(''Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.'') </span> | ||
| − | + | ==Additional Information== | |
| − | + | <br /> | |
| + | ==Links== | ||
| − | '' | + | Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page <span style="color:#0070C0">(''Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "<nowiki>http://www</nowiki>." portion.'')</span> |
| − | + | ==References== | |
| + | (use the "Cite" icon at the top of the page) <span style="color:#0070C0">(''Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted.''</span> <span style="color:#0070C0">''If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference''</span><span style="color:#0070C0">''.''</span><span style="color:#0070C0">) </span> <references /> | ||
| + | '''EXAMPLE Book''' | ||
| − | + | #Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p129-171. | |
| − | + | ==Notes== | |
| − | + | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. | |
| − | + | <nowiki>*</nowiki>''Citation of this Page'': “Acute myeloid leukaemia with NUP98 rearrangement”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_NUP98_rearrangement</nowiki>. | |
| − | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the | ||
| − | |||
| − | |||
| − | < | ||
[[Category:HAEM5]] | [[Category:HAEM5]] | ||
[[Category:DISEASE]] | [[Category:DISEASE]] | ||
[[Category:Diseases A]] | [[Category:Diseases A]] | ||
Latest revision as of 15:30, 29 November 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Eric McGinnis, MD
Fatma Albulushi, MD
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Acute myeloid leukaemia |
| Type | Acute myeloid leukaemia with defining genetic abnormalities |
| Subtype(s) | Acute myeloid leukaemia with NUP98 rearrangement |
WHO Essential and Desirable Genetic Diagnostic Criteria.
| WHO Essential Criteria (Genetics)* | Detection of NUP98 rearrangement |
| WHO Desirable Criteria (Genetics)* | Identification of the NUP98 fusion partner |
| Other Classification | Myeloid blast count may <20% |
*Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the WHO Classification of Tumours.
Definition / Description of Disease
Put your text here (Instructions: Brief description of approximately one paragraph - include disease context relative to other WHO classification categories, diagnostic criteria if applicable, and differential diagnosis if applicable. Other classifications can be referenced for comparison.)
Synonyms / Terminology
Put your text here (Instructions: Include currently used terms and major historical ones, adding “(historical)” after the latter.)
Epidemiology / Prevalence
Put your text here
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
Sites of Involvement
Put your text here (Instruction: Indicate physical sites; EXAMPLE: nodal, extranodal, bone marrow)
Morphologic Features
Put your text here
Immunophenotype
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Finding | Marker |
|---|---|
| Positive (universal) | EXAMPLE: CD1 |
| Positive (subset) | EXAMPLE: CD2 |
| Negative (universal) | EXAMPLE: CD3 |
| Negative (subset) | EXAMPLE: CD4 |
Chromosomal Rearrangements (Gene Fusions)
Acute myeloid leukaemia (AML) with NUP98 rearrangement is characterized by chromosomal translocations involving NUP98 (nucleoporin 98 kDa) on chromosome 11p15.4 and various partner genes. (Reference WHO book). There are over 40 fusion partners which have been reported to date. NUP98 fusions can be categorized into three broad parts. The first category includes NUP98 fusions with transcription factors as partners, which can change the expression of target genes through DNA binding domains. The second category is NUP98 fusions with epigenetic modifiers that modify chromatin to change target gene expression. The third category of NUP98 fusions has neither the DNA binding nor chromatin remodeling domain.[1]
The NUP98 gene (chromosome 11p15) encodes a nucleoporin protein, which is part of the nuclear pore complex which regulates nucleocytoplasmic transport of protein and RNA. NUP98 fusion proteins involve the N-terminal portion of NUP98 and the C-terminal portion of the fusion partner. These fusion partners consist of homeodomain proteins, which are transcription factors, and non-homeodomain proteins, which are thought to play a role in transcriptional or epigenetic regulation.[1][2]
| Driver Gene | Fusion(s) and Common Partner Genes | Molecular Pathogenesis | Typical Chromosomal Alteration(s) | Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
| NUP98 | NUP98::NSD1
|
NUP98-NSD1 prevents EZH2-mediated repression of Hox-A locus genes by colocalizing H3K36 methylation and histone acetylation at regulatory DNA elements hence preventing myeloid progenitor immortalization. | t(5;11)(q35;p15)
Usually cryptic |
Rare (AML) | Defining genetic abnormality in AML | Yes (WHO) | Rare but recurrent alteration seen mainly in children and young adults with AML. Poor overall survival, disease free survival, induction failure and chemotherapy resistance.[2] |
| NUP98 | NUP98::KDM5A | KDM5A is an epigenetic-modifying partners of NUP98 which dysregulate Hox genes expression through recognition of H3K4me3/2 marks by the plant homeodomain (PHD) finger domain. | t(11;12)(p15;p13)
Usually cryptic |
Rare (AML) | Defining genetic abnormality in AML | Yes (WHO) | Commonly associated with erythroid and megakaryocytic phenotypes in pediatric AML (acute erythroid leukemia and acute megakaryocytic leukemia).[2]
Usually associate with unfavorable outcomes |
| NUP98 | NUP98::HOXA9
|
NUP98 fusions bind near the HOX genes loci and activate their expression through chromatin remodeling. The overexpression of distal HoxA cluster genes promote self-renewal and drive leukogenesis. | t(7;11)(p15, p15) | Rare (AML) | Defining genetic abnormality in AML |
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
Individual Region Genomic Gain / Loss / LOH
No characteristic chromosomal gain or loss. However, trisomy 8 and chromosome 13 abnormalities may be observed.
Several reports indicated that del(13q) is a frequent event in NUP98::KDM5A AML patients, indicating co-occurrence of NUP98-KDMA fusion with RB1 deletion.
| Chromosome Number | Gain/Loss/Amp/LOH | Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size] | Relevant Gene(s) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
| 8 | Gain | Trisomy 8 | Unknown | NA | No | |
| 13 | loss | Deletion of 13q | RB1 gene | NA | Particularly associated with NUP98::KDM5A |
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
Gene Mutations (SNV / INDEL)
FLT3-ITD and WT1 mutation are recurring events in NUP98::NSD1 and was also observed in some NUP98::HOXA9 AML patients.(R1). Loss of RB1 at 13q14 is particularly associated with NUP98::KDM5A
| Gene | Genetic Alteration | Tumor Suppressor Gene (TSG)/Oncogene/Other | Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
| FLT3-ITD | Recurrent | Poor prognosis | Seen in 67 to 91% of cases with NUP98::NSD1 | |||
| WT1 | Rare | Reported in 33-55% of NUP98::NSD1 rearranged AML | ||||
| RB1 | Rare | Particularly associated with NUP98::KDM5A |
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
Epigenomic Alterations
Put your text here
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
Genetic Diagnostic Testing Methods
Rearrangements involving NUP98 are often cryptic on conventional karyotype, owing to terminal location of NUP98 on chromosome 11p15.4. Most patients have a normal karyotype. Diagnosis is established using the following tests:
- FISH using NUP98 break-apart probes
- RT-PCR for fusion proteins like NUP98::NSD1
- RNA sequencing
- Optical Genome Mapping (OGM)
Familial Forms
Put your text here (Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.)
Additional Information
Links
Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page (Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "http://www." portion.)
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 Mohanty, Sagarajit (2023-09). "NUP98 Rearrangements in AML: Molecular Mechanisms and Clinical Implications". Onco. 3 (3): 147–164. doi:10.3390/onco3030011. ISSN 2673-7523. Check date values in:
|date=(help) - ↑ 2.0 2.1 2.2 Bertrums, Eline J. M.; et al. (2023-02-23). "Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia". Haematologica. 108 (8): 2044–2058. doi:10.3324/haematol.2022.281653. ISSN 1592-8721.
EXAMPLE Book
- Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p129-171.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Acute myeloid leukaemia with NUP98 rearrangement”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 11/29/2024, https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_NUP98_rearrangement.