Difference between revisions of "Chromophobe renal cell carcinoma"
| [unchecked revision] | [unchecked revision] |
| (18 intermediate revisions by 3 users not shown) | |||
| Line 6: | Line 6: | ||
== Contributors == | == Contributors == | ||
| − | Daynna Wolff PhD FACMG | + | Daynna Wolff, PhD FACMG |
| + | <br> | ||
Yajuan Liu, PhD | Yajuan Liu, PhD | ||
| + | <br> | ||
Rajyasree Emmadi, MD | Rajyasree Emmadi, MD | ||
| + | <br> | ||
Banumathy Gowrishankar, PhD | Banumathy Gowrishankar, PhD | ||
| + | <br> | ||
Jane Houldsworth, PhD | Jane Houldsworth, PhD | ||
| Line 19: | Line 23: | ||
== Description == | == Description == | ||
| − | Chromophobe Renal Cell Carcinoma derives from the intercalated cells of the collecting duct epithelium and accounts for ~5% of renal tumors | + | Chromophobe Renal Cell Carcinoma derives from the intercalated cells of the collecting duct epithelium and accounts for ~5% of renal tumors.<ref>Diaz JI, Mora LB, Hakam A. The Mainz Classification of Renal Cell Tumors. Cancer Control. 1999 Nov;6(6):571-579</ref> |
== IHC Markers == | == IHC Markers == | ||
| Line 33: | Line 37: | ||
! Chromosome !! Gain/Loss/Amp !! Region | ! Chromosome !! Gain/Loss/Amp !! Region | ||
|- | |- | ||
| − | |1 || Loss || Chr1 | + | |1 || Loss || Chr1 (90%) |
|- | |- | ||
| − | |2 || Loss || Chr2 | + | |2 || Loss || Chr2 (80%) |
|- | |- | ||
| − | | | + | |3 || Loss || Chr3 (25%) |
|- | |- | ||
| − | | | + | |5 || Loss || Chr5 (20%) |
|- | |- | ||
| − | | | + | |6 || Loss || Chr6 (90%) |
|- | |- | ||
| − | | | + | |8 || Loss || Chr8 (15%) |
|- | |- | ||
| − | |21 || Loss || Chr21 | + | |9 || Loss || Chr9 (25%) |
| + | |- | ||
| + | |10 || Loss || Chr10 (90%) | ||
| + | |- | ||
| + | |11 || Loss || Chr11 (10%) | ||
| + | |- | ||
| + | |13 || Loss || Chr13 (85%) | ||
| + | |- | ||
| + | |17 || Loss || Chr17 (90%) | ||
| + | |- | ||
| + | |18 || Loss || Chr18 (15%) | ||
| + | |- | ||
| + | |21 || Loss || Chr21 (70%) | ||
|} | |} | ||
| + | |||
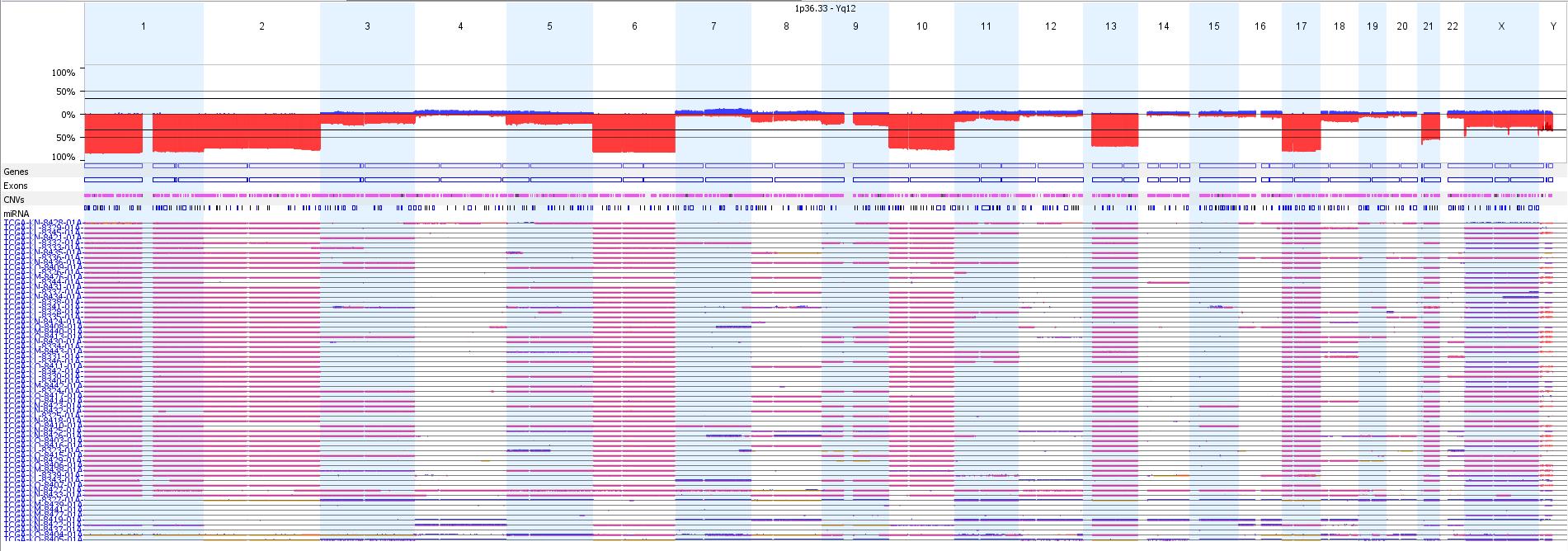
| + | [[File:TCGA-chromophobe RCC.jpeg|center|900px|TCGA Chromophobe RCC copy number profile]] | ||
== Rearrangements == | == Rearrangements == | ||
| − | [[TERT]] (upstream) (5p15) (12%) | + | [[TERT]] (upstream) (5p15) (12%)<ref name=davis>Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, Hacker KE, Bhanot G, Gordenin DA, Chu A, Gunaratne PH, Biehl M, Seth S, Kaipparettu BA, Bristow CA, Donehower LA, Wallen EM, Smith AB, Tickoo SK, Tamboli P, Reuter V, Schmidt LS, Hsieh JJ, Choueiri TK, Hakimi AA; Cancer Genome Atlas Research Network, Chin L, Meyerson M, Kucherlapati R, Park WY, Robertson AG, Laird PW, Henske EP, Kwiatkowski DJ, Park PJ, Morgan M, Shuch B, Muzny D, Wheeler DA, Linehan WM, Gibbs RA, Rathmell WK, Creighton CJ. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014 Sep 8;26(3):319-30.</ref> |
== Mutations (SNV/INDEL) == | == Mutations (SNV/INDEL) == | ||
| − | === From Cosmic Mutated in >20% === | + | === From Cosmic Mutated in >20%<ref>COSMIC (http://cancer.sanger.ac.uk/cosmic)</ref> === |
=== Mutated in 10-20% === | === Mutated in 10-20% === | ||
| − | + | [[TP53]] | |
=== Mutated in 5-10% === | === Mutated in 5-10% === | ||
| − | + | [[VHL]], [[PTEN]] | |
=== Mutated in 2-5% === | === Mutated in 2-5% === | ||
| − | + | [[KMT2D]], [[KMT2C]], [[TERT]], [[MET]], [[ARID1A]], [[FAAH2]], [[PDHB]], [[PDXDC1]], [[ZNF765]] | |
=== mtDNA === | === mtDNA === | ||
== Epigenomics (methylation) == | == Epigenomics (methylation) == | ||
| + | epigenetic silencing of CDKN2A | ||
== Main Pathways Involved == | == Main Pathways Involved == | ||
| + | MTOR pathway targeted (23% cases), increased expression of genes involved in oxidative phosphorylation | ||
== Diagnosis == | == Diagnosis == | ||
| + | Overall loss of whole chormosomes, in particular of chromosomes 1, 2, 6, 10, 13, 17, and 21, eosinophilic variant is mostly diploid<ref>Speicher MR, Schoell B, du Manoir S, Schröck E, Ried T, Cremer T, Störkel S, Kovacs A, Kovacs G. Specific loss of chromosomes 1, 2, 6, 10, 13, 17, and 21 in chromophobe renal cell carcinomas revealed by comparative genomic hybridization. Am J Pathol. 1994 Aug;145(2):356-64.</ref> | ||
== Prognosis == | == Prognosis == | ||
| + | Overall low risk of tumor progression, metastasis, and disease-specific death | ||
== Therapeutics == | == Therapeutics == | ||
Latest revision as of 11:29, 22 August 2016
Contributors
Daynna Wolff, PhD FACMG
Yajuan Liu, PhD
Rajyasree Emmadi, MD
Banumathy Gowrishankar, PhD
Jane Houldsworth, PhD
Tumor Type
Renal Cell Carcinoma
Tumor Classification
Chromophobe Renal Cell Carcinoma
Description
Chromophobe Renal Cell Carcinoma derives from the intercalated cells of the collecting duct epithelium and accounts for ~5% of renal tumors.[1]
IHC Markers
Positive: CD10 , CD117, E-cadherin, EMA, CK7, PAX8, PAX2, AMACR.
Negative: vimentin, RCC, CA-IX.
Genomic Gain/Loss/LOH
| Chromosome | Gain/Loss/Amp | Region |
|---|---|---|
| 1 | Loss | Chr1 (90%) |
| 2 | Loss | Chr2 (80%) |
| 3 | Loss | Chr3 (25%) |
| 5 | Loss | Chr5 (20%) |
| 6 | Loss | Chr6 (90%) |
| 8 | Loss | Chr8 (15%) |
| 9 | Loss | Chr9 (25%) |
| 10 | Loss | Chr10 (90%) |
| 11 | Loss | Chr11 (10%) |
| 13 | Loss | Chr13 (85%) |
| 17 | Loss | Chr17 (90%) |
| 18 | Loss | Chr18 (15%) |
| 21 | Loss | Chr21 (70%) |
Rearrangements
TERT (upstream) (5p15) (12%)[2]
Mutations (SNV/INDEL)
From Cosmic Mutated in >20%[3]
Mutated in 10-20%
Mutated in 5-10%
Mutated in 2-5%
KMT2D, KMT2C, TERT, MET, ARID1A, FAAH2, PDHB, PDXDC1, ZNF765
mtDNA
Epigenomics (methylation)
epigenetic silencing of CDKN2A
Main Pathways Involved
MTOR pathway targeted (23% cases), increased expression of genes involved in oxidative phosphorylation
Diagnosis
Overall loss of whole chormosomes, in particular of chromosomes 1, 2, 6, 10, 13, 17, and 21, eosinophilic variant is mostly diploid[4]
Prognosis
Overall low risk of tumor progression, metastasis, and disease-specific death
Therapeutics
Familial Forms
Birt-Hogg-Dube syndrome (BHD): FLCN (17p11.2)
Links
References
- ↑ Diaz JI, Mora LB, Hakam A. The Mainz Classification of Renal Cell Tumors. Cancer Control. 1999 Nov;6(6):571-579
- ↑ Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, Hacker KE, Bhanot G, Gordenin DA, Chu A, Gunaratne PH, Biehl M, Seth S, Kaipparettu BA, Bristow CA, Donehower LA, Wallen EM, Smith AB, Tickoo SK, Tamboli P, Reuter V, Schmidt LS, Hsieh JJ, Choueiri TK, Hakimi AA; Cancer Genome Atlas Research Network, Chin L, Meyerson M, Kucherlapati R, Park WY, Robertson AG, Laird PW, Henske EP, Kwiatkowski DJ, Park PJ, Morgan M, Shuch B, Muzny D, Wheeler DA, Linehan WM, Gibbs RA, Rathmell WK, Creighton CJ. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014 Sep 8;26(3):319-30.
- ↑ COSMIC (http://cancer.sanger.ac.uk/cosmic)
- ↑ Speicher MR, Schoell B, du Manoir S, Schröck E, Ried T, Cremer T, Störkel S, Kovacs A, Kovacs G. Specific loss of chromosomes 1, 2, 6, 10, 13, 17, and 21 in chromophobe renal cell carcinomas revealed by comparative genomic hybridization. Am J Pathol. 1994 Aug;145(2):356-64.