Difference between revisions of "HAEM5:T-prolymphocytic leukaemia"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| (47 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:T-prolymphocytic leukaemia}} | {{DISPLAYTITLE:T-prolymphocytic leukaemia}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| + | Parastou Tizro, MD, Celeste C. Eno, PhD, Sumire Kitahara, MD | ||
| − | + | Cedars-Sinai, Los Angeles, CA | |
| − | + | ==WHO Classification of Disease== | |
| − | + | {| class="wikitable" | |
| − | + | !Structure | |
| − | + | !Disease | |
| − | + | |- | |
| − | + | |Book | |
| − | + | | | |
| − | + | |- | |
| − | + | |Category | |
| − | + | | | |
| − | + | |- | |
| − | == | + | |Family |
| − | + | | | |
| − | + | |- | |
| − | + | |Type | |
| − | + | | | |
| − | + | |- | |
| − | + | |Subtype(s) | |
| − | + | | | |
| − | == | + | |} |
| − | + | ==WHO Essential and Desirable Genetic Diagnostic Criteria== | |
| − | + | <span style="color:#0070C0">(''Instructions: The table will have the diagnostic criteria from the WHO book <u>autocompleted</u>; remove any <u>non</u>-genetics related criteria. If applicable, add text about other classification'' ''systems that define this entity and specify how the genetics-related criteria differ.'')</span> | |
| − | + | {| class="wikitable" | |
| − | + | |+ | |
| − | + | |WHO Essential Criteria (Genetics)* | |
| − | + | | | |
| + | |- | ||
| + | |WHO Desirable Criteria (Genetics)* | ||
| + | | | ||
| + | |- | ||
| + | |Other Classification | ||
| + | | | ||
| + | |} | ||
| + | <nowiki>*</nowiki>Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the [https://tumourclassification.iarc.who.int/home <u>WHO Classification of Tumours</u>]. | ||
| + | ==Related Terminology== | ||
| + | <span style="color:#0070C0">(''Instructions: The table will have the related terminology from the WHO <u>autocompleted</u>.)''</span> | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | | | + | |+ |
| − | | | + | |Acceptable |
| − | + | | | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | |Not Recommended |
| − | | | + | | |
| − | |||
| − | |||
|} | |} | ||
| − | == | + | ==Gene Rearrangements== |
| + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.'')</span> | ||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Driver Gene!!Fusion(s) and Common Partner Genes!!Molecular Pathogenesis!!Typical Chromosomal Alteration(s) | ||
| + | !Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | ||
| + | !Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | ||
| + | !Established Clinical Significance Per Guidelines - Yes or No (Source) | ||
| + | !Clinical Relevance Details/Other Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''ABL1''||<span class="blue-text">EXAMPLE:</span> ''BCR::ABL1''||<span class="blue-text">EXAMPLE:</span> The pathogenic derivative is the der(22) resulting in fusion of 5’ BCR and 3’ABL1.||<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2) | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (CML) | ||
| + | |<span class="blue-text">EXAMPLE:</span> D, P, T | ||
| + | |<span class="blue-text">EXAMPLE:</span> Yes (WHO, NCCN) | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). BCR::ABL1 is generally favorable in CML (add reference). | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''CIC'' | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''CIC::DUX4'' | ||
| + | |<span class="blue-text">EXAMPLE:</span> Typically, the last exon of ''CIC'' is fused to ''DUX4''. The fusion breakpoint in ''CIC'' is usually intra-exonic and removes an inhibitory sequence, upregulating ''PEA3'' genes downstream of ''CIC'' including ''ETV1'', ''ETV4'', and ''ETV5''. | ||
| + | |<span class="blue-text">EXAMPLE:</span> t(4;19)(q25;q13) | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (CIC-rearranged sarcoma) | ||
| + | |<span class="blue-text">EXAMPLE:</span> D | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | ''DUX4'' has many homologous genes; an alternate translocation in a minority of cases is t(10;19), but this is usually indistinguishable from t(4;19) by short-read sequencing (add references). | |
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''ALK'' | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''ELM4::ALK'' | ||
| − | |||
| − | + | Other fusion partners include ''KIF5B, NPM1, STRN, TFG, TPM3, CLTC, KLC1'' | |
| + | |<span class="blue-text">EXAMPLE:</span> Fusions result in constitutive activation of the ''ALK'' tyrosine kinase. The most common ''ALK'' fusion is ''EML4::ALK'', with breakpoints in intron 19 of ''ALK''. At the transcript level, a variable (5’) partner gene is fused to 3’ ''ALK'' at exon 20. Rarely, ''ALK'' fusions contain exon 19 due to breakpoints in intron 18. | ||
| + | |<span class="blue-text">EXAMPLE:</span> N/A | ||
| + | |<span class="blue-text">EXAMPLE:</span> Rare (Lung adenocarcinoma) | ||
| + | |<span class="blue-text">EXAMPLE:</span> T | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Both balanced and unbalanced forms are observed by FISH (add references). | |
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | + | |<span class="blue-text">EXAMPLE:</span> ''ABL1'' | |
| + | |<span class="blue-text">EXAMPLE:</span> N/A | ||
| + | |<span class="blue-text">EXAMPLE:</span> Intragenic deletion of exons 2–7 in ''EGFR'' removes the ligand-binding domain, resulting in a constitutively active tyrosine kinase with downstream activation of multiple oncogenic pathways. | ||
| + | |<span class="blue-text">EXAMPLE:</span> N/A | ||
| + | |<span class="blue-text">EXAMPLE:</span> Recurrent (IDH-wildtype Glioblastoma) | ||
| + | |<span class="blue-text">EXAMPLE:</span> D, P, T | ||
| + | | | ||
| + | | | ||
|- | |- | ||
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| + | | | ||
|} | |} | ||
| − | + | Rearrangements involving the TCL1 (T-cell leukemia/lymphoma 1) family genes—''TCL1A, MTCP1'' (mature T-cell proliferation), or ''TCL1B'' (also known as ''TCL1/MTCP''1-like 1 [''TML''1])—are highly specific to T-PLL and occur in more than 90% of cases. These translocations juxtapose the ''TRA'' locus with the oncogenes ''TCL1A'' or ''TCL1B'', or in the case of t(X;14), with the ''MTCP1'' gene.<ref name=":6" /><ref name=":7">Matutes E, et al., (2017). T-cell prolymphocytic leukemia, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. Revised 4th Edition. IARC Press: Lyon, France, p346-347.</ref> | |
| − | |||
| − | |||
| − | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| Line 88: | Line 120: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | | | + | |inv(14)(q11.2q32.1) |
| − | + | t(14;14)(q11.2;q32.1) | |
| + | |''TCL1A/B ,TRD''|| ||inv(14) ~60% | ||
| + | t(14;14) ~25% | ||
|Yes | |Yes | ||
|No | |No | ||
| + | |No | ||
| + | |These genetic abnormalities serve as diagnostic markers and generally indicate an aggressive disease. This is due to their role in overexpressing oncogenes like ''TCL1A''. '''Major diagnostic criteria'''.<ref name=":6" /> | ||
| + | |- | ||
| + | |t(X;14)(q28;q11.2) | ||
| + | |''MTCP1, TRD'' | ||
| + | | | ||
| + | |Low (5%) | ||
|Yes | |Yes | ||
| − | | | + | |No |
| + | |No | ||
| + | |'''Major diagnostic criteria'''.<ref name=":6" /> | ||
| + | |} | ||
| − | + | ==Individual Region Genomic Gain/Loss/LOH== | |
| − | | | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene rearrangements. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Can refer to CGC workgroup tables as linked on the homepage if applicable. Please include references throughout the table. Do not delete the table.'') </span> |
| − | + | {| class="wikitable sortable" | |
| − | == | + | |- |
| + | !Chr #!!'''Gain, Loss, Amp, LOH'''!!'''Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size]'''!!'''Relevant Gene(s)''' | ||
| + | !'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' | ||
| + | !'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
| + | !'''Clinical Relevance Details/Other Notes''' | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | 7 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Loss | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | chr7 | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Unknown | ||
| + | |<span class="blue-text">EXAMPLE:</span> D, P | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add references). | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | 8 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Gain | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | chr8 | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Unknown | ||
| + | |<span class="blue-text">EXAMPLE:</span> D, P | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Common recurrent secondary finding for t(8;21) (add references). | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | 17 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Amp | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | 17q12; chr17:39,700,064-39,728,658 [hg38; 28.6 kb] | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | ''ERBB2'' | ||
| + | |<span class="blue-text">EXAMPLE:</span> D, P, T | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Amplification of ''ERBB2'' is associated with HER2 overexpression in HER2 positive breast cancer (add references). Add criteria for how amplification is defined. | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |} | ||
| − | + | Approximately 70-80% of T-PLL karyotypes are complex, which is considered minor diagnostic criteria, and usually include 3-5 or more structural aberrations. Common cytogenetic abnormalities include those of chromosome 8, such as idic(8)(p11.2), t(8;8)(p11.2;q12), and trisomy 8q. Other frequent changes are deletions in 12p13 and 22q, gains in 8q24 (MYC), and abnormalities in chromosomes 5p, 6, and 17.<ref name=":5">Elenitoba-Johnson K, et al. T-prolymphocytic leukemia. In: WHO Classification of Tumours Editorial Board. Haematolymphoid tumours [Internet]. Lyon (France): International Agency for Research on Cancer; 2024 [cited 2024 June 12]. (WHO classification of tumors series, 5th ed.; vol. 11). Available from: https://tumourclassification.iarc.who.int/chaptercontent/63/209</ref> | |
| + | Table: A list of clinically significant and/or recurrent CNAs and CN-LOH with potential or strong diagnostic, prognostic and treatment implications in T-PLL are listed below. | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| Line 110: | Line 204: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | | | + | |8 |
| + | |Gain | ||
| + | |idic(8)(p11) | ||
| − | + | t(8;8)(p11;q12) | |
| − | + | ||
| − | | | + | trisomy 8q<br />8q24 (''MYC'') |
| + | |idic(8)(p11.2) | ||
| − | + | t(8;8)(p11.2;q12) | |
| − | |||
| − | + | trisomy 8q<br />8q24 (''MYC'') | |
| + | |Yes | ||
| + | |No | ||
| + | |No | ||
| + | |Recurrent secondary finding (70-80% of cases). '''Minor diagnostic criteria'''.<ref name=":6">{{Cite journal|last=Staber|first=Philipp B.|last2=Herling|first2=Marco|last3=Bellido|first3=Mar|last4=Jacobsen|first4=Eric D.|last5=Davids|first5=Matthew S.|last6=Kadia|first6=Tapan Mahendra|last7=Shustov|first7=Andrei|last8=Tournilhac|first8=Olivier|last9=Bachy|first9=Emmanuel|date=2019-10-03|title=Consensus criteria for diagnosis, staging, and treatment response assessment of T-cell prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/31292114|journal=Blood|volume=134|issue=14|pages=1132–1143|doi=10.1182/blood.2019000402|issn=1528-0020|pmc=7042666|pmid=31292114}}</ref> | ||
| + | |- | ||
| + | |5 | ||
| + | |Abnormality | ||
| + | |5p, 5q <ref>{{Cite journal|last=Tirado|first=Carlos A.|last2=Starshak|first2=Phillip|last3=Delgado|first3=Paul|last4=Rao|first4=Nagesh|date=2012-08-20|title="T-cell prolymphocytic leukemia (T-PLL), a heterogeneous disease exemplified by two cases and the important role of cytogenetics: a multidisciplinary approach"|url=https://pubmed.ncbi.nlm.nih.gov/23211026|journal=Experimental Hematology & Oncology|volume=1|issue=1|pages=21|doi=10.1186/2162-3619-1-21|issn=2162-3619|pmc=3514161|pmid=23211026}}</ref> | ||
| + | | | ||
| + | |Yes | ||
| + | |Yes | ||
| + | |No | ||
| + | |'''Minor diagnostic criteria'''.<ref name=":6" /> | ||
| + | |- | ||
| + | |6 | ||
| + | |Abnormality | ||
| + | |gain of 6p, loss of 6q <ref>{{Cite journal|last=Dearden|first=Claire|date=2012-07-19|title=How I treat prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/22649104|journal=Blood|volume=120|issue=3|pages=538–551|doi=10.1182/blood-2012-01-380139|issn=1528-0020|pmid=22649104}}</ref><ref>{{Cite journal|last=Soulier|first=J.|last2=Pierron|first2=G.|last3=Vecchione|first3=D.|last4=Garand|first4=R.|last5=Brizard|first5=F.|last6=Sigaux|first6=F.|last7=Stern|first7=M. H.|last8=Aurias|first8=A.|date=2001-07|title=A complex pattern of recurrent chromosomal losses and gains in T-cell prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/11391795|journal=Genes, Chromosomes & Cancer|volume=31|issue=3|pages=248–254|doi=10.1002/gcc.1141|issn=1045-2257|pmid=11391795}}</ref> | ||
| + | | | ||
| + | |No | ||
| + | |No | ||
| + | |No | ||
| + | | | ||
| + | |- | ||
| + | |11 | ||
| + | |Loss | ||
| + | |11q | ||
| + | |ch11q21-q23.3 | ||
| + | |Yes | ||
| + | |Yes | ||
| + | |Yes (poor) | ||
| + | |Frequent, '''Minor diagnostic criteria'''.<ref name=":6" /> | ||
| + | |- | ||
| + | |12 | ||
| + | |Loss | ||
| + | |12p | ||
| + | |12p13 | ||
|Yes | |Yes | ||
|Yes | |Yes | ||
|No | |No | ||
| − | | | + | |Haploinsufficiency of the ''CDKN1B'' gene at the 12p13 locus contributes to the development of T-PLL.<ref>{{Cite journal|last=Le Toriellec|first=Emilie|last2=Despouy|first2=Gilles|last3=Pierron|first3=Gaëlle|last4=Gaye|first4=Nogaye|last5=Joiner|first5=Marjorie|last6=Bellanger|first6=Dorine|last7=Vincent-Salomon|first7=Anne|last8=Stern|first8=Marc-Henri|date=2008-02-15|title=Haploinsufficiency of CDKN1B contributes to leukemogenesis in T-cell prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/18073348|journal=Blood|volume=111|issue=4|pages=2321–2328|doi=10.1182/blood-2007-06-095570|issn=0006-4971|pmid=18073348}}</ref> |
| − | + | '''Minor diagnostic criteria'''.<ref name=":6" /> | |
| − | |||
|- | |- | ||
| − | | | + | |13 |
| − | + | |Loss | |
| − | + | |13q | |
| − | | | + | |13q14.3 |
| − | | | + | |Yes |
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | |||
|No | |No | ||
|No | |No | ||
| + | |'''Minor diagnostic criteria'''.<ref name=":6" /> | ||
| + | |- | ||
| + | |17 | ||
| + | |Loss | ||
| + | |17p | ||
| + | |17p13 | ||
|No | |No | ||
| − | | | + | |Yes |
| + | |Yes (resistance to therapy) | ||
| + | |May include ''TP53'' gene at 17p13.1 <ref name=":7" /> | ||
| + | |- | ||
| + | |22 | ||
| + | |Loss | ||
| + | |Monosomy 22 | ||
| + | del(22q) | ||
| + | | | ||
| + | 22q11-12 <ref>{{Cite journal|last=Stengel|first=Anna|last2=Kern|first2=Wolfgang|last3=Zenger|first3=Melanie|last4=Perglerová|first4=Karolina|last5=Schnittger|first5=Susanne|last6=Haferlach|first6=Torsten|last7=Haferlach|first7=Claudia|date=2014-12-06|title=A Comprehensive Cytogenetic and Molecular Genetic Characterization of Patients with T-PLL Revealed Two Distinct Genetic Subgroups and JAK3 Mutations As an Important Prognostic Marker|url=https://doi.org/10.1182/blood.V124.21.1639.1639|journal=Blood|volume=124|issue=21|pages=1639–1639|doi=10.1182/blood.v124.21.1639.1639|issn=0006-4971}}</ref><ref name=":0">{{Cite journal|last=Fang|first=Hong|last2=Beird|first2=Hannah C.|last3=Wang|first3=Sa A.|last4=Ibrahim|first4=Andrew F.|last5=Tang|first5=Zhenya|last6=Tang|first6=Guilin|last7=You|first7=M. James|last8=Hu|first8=Shimin|last9=Xu|first9=Jie|date=2023-09|title=T-prolymphocytic leukemia: TCL1 or MTCP1 rearrangement is not mandatory to establish diagnosis|url=https://pubmed.ncbi.nlm.nih.gov/37443196|journal=Leukemia|volume=37|issue=9|pages=1919–1921|doi=10.1038/s41375-023-01956-3|issn=1476-5551|pmid=37443196}}</ref> | ||
| − | + | (most common) | |
| + | |Yes | ||
| + | |No | ||
| + | |No | ||
| + | |Leading to the dysregulation of genes such as ''BCL11B'', which is crucial in T-cell development and function.<ref name=":0" /> | ||
| + | '''Minor diagnostic criteria'''.<ref name=":6" /> | ||
|} | |} | ||
| − | |||
| − | Put your text here <span style="color:#0070C0">('' | + | ==Diagnostic criteria== |
| + | Diagnosis requires either <u>all three major criteria</u> '''or''' the <u>first two major criteria along with one minor criterion</u>:<ref name=":5" /> | ||
| + | |||
| + | *'''Major criteria:''' | ||
| + | **5 x 10<sup>9</sup>/L cells of T PLL phenotype in peripheral blood or bone marrow | ||
| + | **T cell clonality by molecular or flow cytometry methods | ||
| + | **Abnormalities of 14q32 or Xq28 or expression of TCL1A/B or MTC** | ||
| + | *'''Minor criteria:''' | ||
| + | **Abnormalities involving chromosome 11 | ||
| + | **Abnormalities in chromosome 8 | ||
| + | **Abnormalities in chromosome 5, 12, 13, 22 or complex karyotype | ||
| + | **Involvement of specific sites (spleen, effusions) | ||
| + | |||
| + | <nowiki>**</nowiki>Cases lacking these abnormalities may be referred to as "TCL1 family-negative T-PLL." by some investigators<ref name=":0" />. It is, however, recommended in WHO-HAEM5 that such cases should preferably be classified as peripheral T-cell lymphoma NOS with leukemic involvement (after exclusion of other specific leukemic T-cell entities) as there are insufficient clinicopathological and molecular data to determine the relationship of TCL1 family–negative T-PLL to T-PLL. | ||
| + | |||
| + | ==Characteristic Chromosomal or Other Global Mutational Patterns== | ||
| + | Put your text here and fill in the table <span style="color:#0070C0">(I''nstructions: Included in this category are alterations such as hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis; microsatellite instability; homologous recombination deficiency; mutational signature pattern; etc. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.'')</span> | ||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Chromosomal Pattern | ||
| + | !Molecular Pathogenesis | ||
| + | !'''Prevalence -''' | ||
| + | '''Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' | ||
| + | !'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' | ||
| + | !'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
| + | !'''Clinical Relevance Details/Other Notes''' | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Co-deletion of 1p and 18q | ||
| + | |<span class="blue-text">EXAMPLE:</span> See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (Oligodendroglioma) | ||
| + | |<span class="blue-text">EXAMPLE:</span> D, P | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Microsatellite instability - hypermutated | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (Endometrial carcinoma) | ||
| + | |<span class="blue-text">EXAMPLE:</span> P, T | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |} | ||
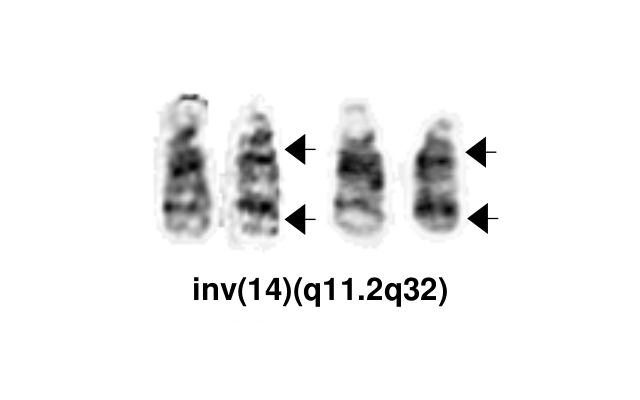
| + | [[File:Inv(14)(q11.2q32).png|thumb|Inv(14)(q11.2q32)]] | ||
| + | The most common chromosomal abnormality in T-PLL involves an inversion of chromosome 14, with breakpoints at q11.2 and q32.1, observed in about 60-80% of patients and described as inv(14). Additionally, in 10-20% of cases, there is a translocation t(14;14)(q11.2;q32.1).<ref name=":5" /> <ref name=":7" /> | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| Line 156: | Line 352: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | | | + | |inv(14)(q11q32) |
| − | + | t(14;14)(q11.2;q32.1) | |
| − | |||
|Yes | |Yes | ||
|No | |No | ||
|No | |No | ||
| − | | | + | |The most common chromosomal abnormality in T-PLL involves an inversion of chromosome 14, with breakpoints at q11.2 and q32.1, observed in about 60-80% of patients and described as inv(14). Additionally, in 10-20% of cases, there is a translocation t(14;14)(q11.2;q32.1) |
| + | |} | ||
| − | + | ==Gene Mutations (SNV/INDEL)== | |
| − | | | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent or common as well either disease defining and/or clinically significant. If a gene has multiple mechanisms depending on the type or site of the alteration, add multiple entries in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Details on clinical significance such as prognosis and other important information such as concomitant and mutually exclusive mutations can be provided in the notes section. Please include references throughout the table. Do not delete the table.'') </span> |
| − | ==Gene | + | {| class="wikitable sortable" |
| + | |- | ||
| + | !Gene!!'''Genetic Alteration'''!!'''Tumor Suppressor Gene, Oncogene, Other'''!!'''Prevalence -''' | ||
| + | '''Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' | ||
| + | !'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T ''' | ||
| + | !'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
| + | !'''Clinical Relevance Details/Other Notes''' | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span>''EGFR'' | ||
| + | |||
| + | <br /> | ||
| + | |<span class="blue-text">EXAMPLE:</span> Exon 18-21 activating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Oncogene | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (lung cancer) | ||
| + | |<span class="blue-text">EXAMPLE:</span> T | ||
| + | |<span class="blue-text">EXAMPLE:</span> Yes (NCCN) | ||
| + | |<span class="blue-text">EXAMPLE:</span> Exons 18, 19, and 21 mutations are targetable for therapy. Exon 20 T790M variants cause resistance to first generation TKI therapy and are targetable by second and third generation TKIs (add references). | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''TP53''; Variable LOF mutations | ||
| + | <br /> | ||
| + | |<span class="blue-text">EXAMPLE:</span> Variable LOF mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Tumor Supressor Gene | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (breast cancer) | ||
| + | |<span class="blue-text">EXAMPLE:</span> P | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> >90% are somatic; rare germline alterations associated with Li-Fraumeni syndrome (add reference). Denotes a poor prognosis in breast cancer. | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''BRAF''; Activating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Activating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Oncogene | ||
| + | |<span class="blue-text">EXAMPLE:</span> Common (melanoma) | ||
| + | |<span class="blue-text">EXAMPLE:</span> T | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |}Note: A more extensive list of mutations can be found in [https://www.cbioportal.org/ <u>cBioportal</u>], [https://cancer.sanger.ac.uk/cosmic <u>COSMIC</u>], and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | ||
| − | + | Although gene mutations beyond ''TCL1'' family alterations are not yet recognized as diagnostic criteria and remain under investigation for T-PLL, the mutational landscape of T-PLL provides valuable insights. These discoveries open up potential avenues for novel targeted therapies in treating this aggressive form of leukemia. | |
| + | Deletions and mutations of the ATM gene (present in up to 90% of T-PLL cases but typically absent in other mature T-cell malignancies) are considered highly indicative for a diagnosis of suspected TCL1 family-negative T-PLL.<ref name=":8">{{Cite journal|last=Schrader|first=A.|last2=Crispatzu|first2=G.|last3=Oberbeck|first3=S.|last4=Mayer|first4=P.|last5=Pützer|first5=S.|last6=von Jan|first6=J.|last7=Vasyutina|first7=E.|last8=Warner|first8=K.|last9=Weit|first9=N.|date=2018-02-15|title=Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL|url=https://pubmed.ncbi.nlm.nih.gov/29449575|journal=Nature Communications|volume=9|issue=1|pages=697|doi=10.1038/s41467-017-02688-6|issn=2041-1723|pmc=5814445|pmid=29449575}}</ref><ref name=":3" /> | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| Line 178: | Line 417: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | | | + | |''ATM'' |
| + | |TSG | ||
| + | |53% (COSMIC) | ||
| + | |''ATM'' mutation/deletion | ||
| + | |None specified | ||
| + | |Yes | ||
| + | |Yes | ||
| + | |Yes (PARP inhibitors, NCT03263637) | ||
| + | |Since deletions of or missense mutations at the ''ATM'' locus are found in up to 80% to 90% of T-PLL cases, ''ATM'' alterations can serve as a minor diagnostic criterion.<ref name=":6" /><ref name=":8" /> | ||
| + | |- | ||
| + | |''FBXW10'' | ||
| + | |TSG | ||
| + | |72% (COSMIC) | ||
| + | |''JAK/STAT'' pathway | ||
| + | |None specified | ||
| + | |Unknown | ||
| + | |Unknown | ||
| + | |Unknown | ||
| + | | | ||
| + | |- | ||
| + | |''IL2RG,'' ''JAK1, JAK3, STAT5B'' | ||
| + | |Oncogene | ||
| + | |8% ''JAK1'' | ||
| + | |||
| + | 34% ''JAK3'' | ||
| − | + | 16% ''STAT5B'' | |
| − | + | 2% ''IL2RG'' | |
| − | + | (COSMIC) | |
| − | |||
| − | |||
| − | + | (cumulative prevalence of ~ 60%)<ref>{{Cite journal|last=Wahnschaffe|first=Linus|last2=Braun|first2=Till|last3=Timonen|first3=Sanna|last4=Giri|first4=Anil K.|last5=Schrader|first5=Alexandra|last6=Wagle|first6=Prerana|last7=Almusa|first7=Henrikki|last8=Johansson|first8=Patricia|last9=Bellanger|first9=Dorine|date=2019-11-21|title=JAK/STAT-Activating Genomic Alterations Are a Hallmark of T-PLL|url=https://pubmed.ncbi.nlm.nih.gov/31766351|journal=Cancers|volume=11|issue=12|pages=1833|doi=10.3390/cancers11121833|issn=2072-6694|pmc=6966610|pmid=31766351}}</ref> | |
| − | | | + | |''ATM, TP53'', Epigenetic modifiers <ref name=":1">{{Cite journal|last=Andersson|first=E. I.|last2=Pützer|first2=S.|last3=Yadav|first3=B.|last4=Dufva|first4=O.|last5=Khan|first5=S.|last6=He|first6=L.|last7=Sellner|first7=L.|last8=Schrader|first8=A.|last9=Crispatzu|first9=G.|date=2018-03|title=Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling|url=https://pubmed.ncbi.nlm.nih.gov/28804127|journal=Leukemia|volume=32|issue=3|pages=774–787|doi=10.1038/leu.2017.252|issn=1476-5551|pmid=28804127}}</ref><ref name=":2">{{Cite journal|last=Pinter-Brown|first=Lauren C.|date=2021-12-30|title=JAK/STAT: a pathway through the maze of PTCL?|url=https://doi.org/10.1182/blood.2021014238|journal=Blood|volume=138|issue=26|pages=2747–2748|doi=10.1182/blood.2021014238|issn=0006-4971}}</ref> |
| − | | | + | |Typically, mutations within this pathway occur in a mutually exclusive manner.<ref name=":3">{{Cite journal|last=Kiel|first=Mark J.|last2=Velusamy|first2=Thirunavukkarasu|last3=Rolland|first3=Delphine|last4=Sahasrabuddhe|first4=Anagh A.|last5=Chung|first5=Fuzon|last6=Bailey|first6=Nathanael G.|last7=Schrader|first7=Alexandra|last8=Li|first8=Bo|last9=Li|first9=Jun Z.|date=2014-08-28|title=Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/24825865|journal=Blood|volume=124|issue=9|pages=1460–1472|doi=10.1182/blood-2014-03-559542|issn=1528-0020|pmc=4148768|pmid=24825865}}</ref> |
| − | | | + | |Yes |
| − | | | + | |Yes |
| − | | | + | |Yes |
| − | | | + | |Targeting this pathway with specific ''JAK/STAT'' pathway inhibitors, such as tofacitinib, has shown promise in preclinical studies and early clinical trials. Combining JAK/STAT inhibitors with other treatments, like BCL-2 inhibitors, may enhance therapeutic efficacy and improve outcomes for T-PLL patients<ref>{{Cite journal|last=Gomez-Arteaga|first=Alexandra|last2=Margolskee|first2=Elizabeth|last3=Wei|first3=Mike T.|last4=van Besien|first4=Koen|last5=Inghirami|first5=Giorgio|last6=Horwitz|first6=Steven|date=2019-07|title=Combined use of tofacitinib (pan-JAK inhibitor) and ruxolitinib (a JAK1/2 inhibitor) for refractory T-cell prolymphocytic leukemia (T-PLL) with a JAK3 mutation|url=https://pubmed.ncbi.nlm.nih.gov/30997845|journal=Leukemia & Lymphoma|volume=60|issue=7|pages=1626–1631|doi=10.1080/10428194.2019.1594220|issn=1029-2403|pmc=8162842|pmid=30997845}}</ref><ref>{{Cite journal|url=https://ashpublications.org/blood/article/126/23/5486/134544/Refractory-TCell-Prolymphocytic-Leukemia-with-JAK3|doi=10.1182/blood.v126.23.5486.5486}}</ref> |
| − | < | + | |- |
| − | |} | + | |''EZH2'' |
| − | Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | + | |Both oncogene and TSG |
| + | |16% (COSMIC) | ||
| + | |''JAK/STAT'' pathway<ref name=":1" /><ref name=":2" /> | ||
| + | |None specified | ||
| + | |No | ||
| + | |No (see note) | ||
| + | |See note | ||
| + | |''EZH2'' inhibitors like tazemetostat have shown efficacy in other hematologic malignancies, providing a rationale for their potential use in T-PLL | ||
| + | |- | ||
| + | |''BCOR'' | ||
| + | |TSG | ||
| + | |8% (COSMIC) | ||
| + | |''JAK/STAT'' pathway<ref name=":1" /><ref name=":2" /> | ||
| + | |None specified | ||
| + | |No | ||
| + | |No (see note) | ||
| + | |No | ||
| + | |A negative impact on overall survival (OS) was not observed for T-PLL patients in the study. However, this might be attributable to the relatively low number of cases compared to studies on AML and MDS.<ref name=":9">{{Cite journal|last=Stengel|first=Anna|last2=Kern|first2=Wolfgang|last3=Zenger|first3=Melanie|last4=Perglerová|first4=Karolína|last5=Schnittger|first5=Susanne|last6=Haferlach|first6=Torsten|last7=Haferlach|first7=Claudia|date=2016-01|title=Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker|url=https://pubmed.ncbi.nlm.nih.gov/26493028|journal=Genes, Chromosomes & Cancer|volume=55|issue=1|pages=82–94|doi=10.1002/gcc.22313|issn=1098-2264|pmid=26493028}}</ref> | ||
| + | |- | ||
| + | |''SAMHD1'' | ||
| + | |TSG | ||
| + | |~7-20%<ref name=":8" /><ref name=":4">{{Cite journal|last=Johansson|first=Patricia|last2=Klein-Hitpass|first2=Ludger|last3=Choidas|first3=Axel|last4=Habenberger|first4=Peter|last5=Mahboubi|first5=Bijan|last6=Kim|first6=Baek|last7=Bergmann|first7=Anke|last8=Scholtysik|first8=René|last9=Brauser|first9=Martina|date=2018-01-19|title=SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/29352181|journal=Blood Cancer Journal|volume=8|issue=1|pages=11|doi=10.1038/s41408-017-0036-5|issn=2044-5385|pmc=5802577|pmid=29352181}}</ref> | ||
| + | |None specified | ||
| + | |None specified | ||
| + | |Yes | ||
| + | |Yes | ||
| + | |No | ||
| + | |''SAMHD1'' mutations may indicate a defective DNA damage response and aggressive disease <ref name=":4" /> | ||
| + | |- | ||
| + | |''CHEK2'' | ||
| + | |TSG | ||
| + | |5% (COSMIC) | ||
| + | |''ATM, TP53, JAK/STA''T pathway, Epigenetic modifiers | ||
| + | |None specified | ||
| + | |No | ||
| + | |Yes | ||
| + | |No | ||
| + | |''CHEK2'' mutations may indicate a defective DNA damage response and aggressive disease <ref name=":3" /><ref>{{Cite journal|last=Braun|first=Till|last2=Dechow|first2=Annika|last3=Friedrich|first3=Gregor|last4=Seifert|first4=Michael|last5=Stachelscheid|first5=Johanna|last6=Herling|first6=Marco|date=2021|title=Advanced Pathogenetic Concepts in T-Cell Prolymphocytic Leukemia and Their Translational Impact|url=https://pubmed.ncbi.nlm.nih.gov/34869023|journal=Frontiers in Oncology|volume=11|pages=775363|doi=10.3389/fonc.2021.775363|issn=2234-943X|pmc=8639578|pmid=34869023}}</ref> | ||
| + | |- | ||
| + | |''TP53'' | ||
| + | |TSG | ||
| + | |2% (COSMIC) | ||
| + | |''ATM, JAK/STA''T pathway, Epigenetic modifiers | ||
| + | |None specified-In a study of T-PLL case, TP53 mutations were predominantly found in patients lacking TCRA/D rearrangements.<ref name=":9" /> | ||
| + | |No | ||
| + | |Yes | ||
| + | |Associated with resistance to therapy | ||
| + | |Mutations in TP53 are less frequent than deletions.<ref name=":9" />May show overexpression of p53 in some cases.<ref name=":7" /> | ||
| + | |}Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | ||
==Epigenomic Alterations== | ==Epigenomic Alterations== | ||
| − | + | Research indicates that epigenetic modifications in the regulatory regions of key oncogenes and genes involved in DNA damage response and T-cell receptor regulation are clearly present. These changes are closely associated with the transcriptional dysregulation that forms the core lesions of T-PLL.<ref>{{Cite journal|last=Tian|first=Shulan|last2=Zhang|first2=Henan|last3=Zhang|first3=Pan|last4=Kalmbach|first4=Michael|last5=Lee|first5=Jeong-Heon|last6=Ordog|first6=Tamas|last7=Hampel|first7=Paul J.|last8=Call|first8=Timothy G.|last9=Witzig|first9=Thomas E.|date=2021-04-15|title=Epigenetic alteration contributes to the transcriptional reprogramming in T-cell prolymphocytic leukemia|url=https://pubmed.ncbi.nlm.nih.gov/33859327|journal=Scientific Reports|volume=11|issue=1|pages=8318|doi=10.1038/s41598-021-87890-9|issn=2045-2322|pmc=8050249|pmid=33859327}}</ref> | |
| − | |||
| − | |||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | + | The key pathways involved in the pathogenesis of T-cell prolymphocytic leukemia (T-PLL) include DNA damage repair, T-cell receptor (''TCR'') signaling, and epigenetic modulation. Additionally, there is frequent mutational activation of the ''IL2RG-JAK1-JAK3-STAT5B'' pathway, which plays a significant role in the disease's development and progression.<ref name=":6" /> | |
| − | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | | | + | |''TCL1A/B rearrangement'' |
| − | | | + | |''AKT'' signaling and TCR signal amplification pathways |
| − | | | + | |Increased cell survival and proliferation |
|- | |- | ||
| − | | | + | |''MTCP1'' |
| − | | | + | |''AKT'' signaling and TCR signal amplification pathways |
| − | | | + | |Increased cell survival and proliferation |
|- | |- | ||
| − | | | + | |''ATM, CHEK2'' |
| − | | | + | |DNA damage repair pathway |
| − | | | + | |Genomic instability |
| + | |- | ||
| + | |''JAK1, JAK3, STAT5B'' | ||
| + | |''JAK-STAT'' pathway | ||
| + | |Unchecked cell growth and survival | ||
| + | |- | ||
| + | |''IL2RG'' | ||
| + | |''JAK-STAT'' pathway, Cytokine signaling | ||
| + | |Promoting lymphocyte proliferation | ||
| + | |- | ||
| + | |''EZH2'' | ||
| + | |Transcription regulator | ||
| + | |Altering the epigenetic landscape | ||
|} | |} | ||
==Genetic Diagnostic Testing Methods== | ==Genetic Diagnostic Testing Methods== | ||
| + | Diagnosing T-PLL involves a combination of clinical evaluation, laboratory tests, imaging studies, and genetic testing to identify diagnostic criteria. T-cell clonality can be confirmed through PCR, NGS, or flow cytometry.<ref>{{Cite journal|last=Kotrova|first=Michaela|last2=Novakova|first2=Michaela|last3=Oberbeck|first3=Sebastian|last4=Mayer|first4=Petra|last5=Schrader|first5=Alexandra|last6=Knecht|first6=Henrik|last7=Hrusak|first7=Ondrej|last8=Herling|first8=Marco|last9=Brüggemann|first9=Monika|date=2018-11|title=Next-generation amplicon TRB locus sequencing can overcome limitations of flow-cytometric Vβ expression analysis and confirms clonality in all T-cell prolymphocytic leukemia cases|url=https://pubmed.ncbi.nlm.nih.gov/30414304|journal=Cytometry. Part A: The Journal of the International Society for Analytical Cytology|volume=93|issue=11|pages=1118–1124|doi=10.1002/cyto.a.23604|issn=1552-4930|pmid=30414304}}</ref> Patients with T-PLL often exhibit complex karyotypes with recurrent genetic features that aid in diagnosis. Therefore, cytogenetic studies are useful for distinguishing T-PLL from other T-lymphoproliferative disorders.<ref name=":6" /> | ||
| + | |||
| + | *'''Cytogenetic Analysis''' | ||
| + | |||
| + | #Karyotyping: To identify characteristic chromosomal abnormalities, such as inv(14)(q11q32), t(14;14)(q11;q32), or other translocations involving chromosome 14. '''Major diagnostic criteria''' | ||
| + | #Fluorescence In Situ Hybridization (FISH): To detect specific genetic abnormalities, such as TCL1 gene rearrangements as a '''Major diagnostic criterion''' or MYC as a '''Minor diagnostic criterion''' (alternatively, molecular studies could be used). see note. | ||
| + | |||
| + | <small><u>'''Note:''' ''TCL1A'' break-apart probes specific to the 14q32 region can identify translocations involving TCL1A. When a ''TCL1A'' rearrangement is not identified and the patient has T-cell prolymphocytic leukemia/lymphoma (T-PLL), reflex testing using the ''TRAD'' break-apart probe set may be performed.</u></small> | ||
| + | |||
| + | *'''Molecular Genetic Testing''' | ||
| − | + | #Polymerase Chain Reaction (PCR) and Reverse Transcription PCR (RT-PCR): To show the rearrangements of the TR gene (TCRB, TCRG loci) as a '''Major diagnostic criterion,''' and alternative to FISH rearrangements of the ''TCL1'' or ''MTCP'' genes at the ''TRD'' locus can be detected by PCR. '''Major diagnostic criteria''' | |
| + | #Next generation sequencing (NGS)-See note | ||
| + | <u>'''<small>Note:</small>''' <small>Although alterations of ''TCL1A'', ''TCL1B (TML1)'', or ''MTCP'' are present in more than 90% of cases, they are not found in 100% of cases. Taken together, assessment of clonal TCR rearrangement, cytogenetics, and FISH are relevant genetic tests to establish the diagnosis of T-PLL. Genetic sequencing is currently not a diagnostic requirement; however, it may provide information regarding the underlying pathogenesis of T-PLL or might help to identify relevant prognostic subgroups.</small></u><ref name=":6" /> | ||
==Familial Forms== | ==Familial Forms== | ||
| + | While there is no noticeable familial clustering of T-cell prolymphocytic leukemia (T-PLL), a subset of cases can develop in the context of ataxia-telangiectasia (AT). AT is characterized by germline mutations in the ''ATM'' gene, and patients with AT are at an increased risk for various malignancies, including T-PLL. In these cases, biallelic inactivation of the ''ATM'' tumor suppressor gene occurs, with about 10% to 15% penetrance of the tumor phenotype by early adulthood. T-PLL represents nearly 3% of all malignancies in patients with ataxia-telangiectasia. <ref>{{Cite journal|last=Suarez|first=Felipe|last2=Mahlaoui|first2=Nizar|last3=Canioni|first3=Danielle|last4=Andriamanga|first4=Chantal|last5=Dubois d'Enghien|first5=Catherine|last6=Brousse|first6=Nicole|last7=Jais|first7=Jean-Philippe|last8=Fischer|first8=Alain|last9=Hermine|first9=Olivier|date=2015-01-10|title=Incidence, presentation, and prognosis of malignancies in ataxia-telangiectasia: a report from the French national registry of primary immune deficiencies|url=https://pubmed.ncbi.nlm.nih.gov/25488969|journal=Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology|volume=33|issue=2|pages=202–208|doi=10.1200/JCO.2014.56.5101|issn=1527-7755|pmid=25488969}}</ref> <ref>{{Cite journal|last=Taylor|first=A. M.|last2=Metcalfe|first2=J. A.|last3=Thick|first3=J.|last4=Mak|first4=Y. F.|date=1996-01-15|title=Leukemia and lymphoma in ataxia telangiectasia|url=https://pubmed.ncbi.nlm.nih.gov/8555463|journal=Blood|volume=87|issue=2|pages=423–438|issn=0006-4971|pmid=8555463}}</ref> <ref>{{Cite journal|last=Li|first=Geling|last2=Waite|first2=Emily|last3=Wolfson|first3=Julie|date=2017-12-26|title=T-cell prolymphocytic leukemia in an adolescent with ataxia-telangiectasia: novel approach with a JAK3 inhibitor (tofacitinib)|url=https://pubmed.ncbi.nlm.nih.gov/29296924|journal=Blood Advances|volume=1|issue=27|pages=2724–2728|doi=10.1182/bloodadvances.2017010470|issn=2473-9529|pmc=5745136|pmid=29296924}}</ref> | ||
| + | ==Additional Information== | ||
| + | |||
| + | * In T-PLL, the rapid growth of the disease necessitates immediate initiation of treatment. The most effective first-line treatment is alemtuzumab, an anti-CD52 antibody with remission rates over 80%. However, these remissions usually last only 1-2 years. To potentially extend remission, eligible patients are advised to undergo allogeneic blood stem cell transplantation (allo-SCT) during their first complete remission, which can lead to longer remission durations of over 4-5 years for 15-30% of patients. Consequently, the prognosis for T-PLL remains poor, with median overall survival times under two years and five-year survival rates below 5%[https://clinicaltrials.gov/study/NCT03989466 . Ongoing studies are exploring molecularly targeted drugs and signaling pathway inhibitors, for routine clinical use in treating T-PLL.] | ||
| + | |||
| + | This disease is <u>defined/characterized</u> as detailed below: | ||
| + | |||
| + | * T-prolymphocytic leukemia (T-PLL) is an aggressive form of T-cell leukemia marked by the proliferation of small to medium-sized prolymphocytes exhibiting a mature post-thymic T-cell phenotype.<ref name=":5" /> | ||
| + | |||
| + | The <u>epidemiology/prevalence</u> of this disease is detailed below: | ||
| + | |||
| + | * T-PLL is an uncommon disease, accounting for approximately 2% of all mature lymphoid leukemias in adults. It mainly affects older individuals, with a median onset age of 65 years, ranging from 30 to 94 years. The disorder exhibits a slight male predominance, with a male to female ratio of 1.33:1.<ref name=":5" /> | ||
| + | |||
| + | The <u>clinical features</u> of this disease are detailed below: | ||
| + | |||
| + | * The most prevalent symptom of the disease is a leukemic presentation, characterized by a rapid, exponential increase in lymphocyte counts, which exceed 100 × 10^9/L in 75% of patients. Approximately 30% of patients may initially experience an asymptomatic, slow-progressing phase, but this typically develops into an active disease state.<ref name=":5" /><ref name=":6" /> | ||
| + | |||
| + | Signs and symptoms - B symptoms (Fever, night sweats, weight loss); Hepatosplenomegaly (Frequently observed); Generalized lymphadenopathy with slightly enlarged lymph nodes (Frequently observed); Cutaneous involvement (20%); Malignant effusions (15%) | ||
| + | |||
| + | Laboratory findings - Anemia and thrombocytopenia; Marked lymphocytosis > 100 × 10^9/L (75% of cases); Atypical lymphocytosis > 5 × 10^9/L; Serum lactate dehydrogenase (LDH) (increased-may reflect disease burden); β2 microglobulin (B2M) (increased-may reflect disease burden) | ||
| + | |||
| + | The <u>sites of involvement</u> of this disease are detailed below: | ||
| + | |||
| + | * Peripheral blood, bone marrow, spleen (mostly red pulp), liver, lymph node (mostly paracortical), and sometimes skin and serosa (primarily pleura). Extra lymphatic and extramedullary atypical manifestations including skin, muscles and intestines are particularly common in relapse.<ref name=":5" /> | ||
| − | + | The <u>morphologic features</u> of this disease are detailed below: | |
| − | == | + | * Blood smears in T-PLL typically reveal anemia, thrombocytopenia, and leukocytosis, with atypical lymphocytes in three morphological forms: The most common form (75% of cases) features medium-sized cells with a high nuclear-to-cytoplasmic ratio, moderately condensed chromatin, a single visible nucleolus, and slightly basophilic cytoplasm. In 20% of cases, the cells appear as a small cell variant with densely condensed chromatin and an inconspicuous nucleolus. About 5% of cases exhibit a cerebriform variant with irregular nuclei resembling those in mycosis fungoides. Regardless of the nuclear features, a common morphological characteristic is the presence of cytoplasmic protrusions or blebs.<ref>{{Cite journal|last=Gutierrez|first=Marc|last2=Bladek|first2=Patrick|last3=Goksu|first3=Busra|last4=Murga-Zamalloa|first4=Carlos|last5=Bixby|first5=Dale|last6=Wilcox|first6=Ryan|date=2023-07-28|title=T-Cell Prolymphocytic Leukemia: Diagnosis, Pathogenesis, and Treatment|url=https://pubmed.ncbi.nlm.nih.gov/37569479|journal=International Journal of Molecular Sciences|volume=24|issue=15|pages=12106|doi=10.3390/ijms241512106|issn=1422-0067|pmc=PMC10419310|pmid=37569479}}</ref>Bone marrow aspirates show clusters of these neoplastic cells, with a mixed pattern of involvement including diffuse and interstitial, in trephine core biopsy.<ref name=":6" /> |
| − | + | The <u>immunophenotype</u> of this disease is detailed below: | |
| − | == | + | * '''Cytochemistry:''' T-cell prolymphocytes show strong staining with alpha-naphthyl acetate esterase and acid phosphatase, presenting a distinctive dot-like pattern, but cytochemistry is not commonly used for diagnosis.<ref>{{Cite journal|last=Yang|first=K.|last2=Bearman|first2=R. M.|last3=Pangalis|first3=G. A.|last4=Zelman|first4=R. J.|last5=Rappaport|first5=H.|date=1982-08|title=Acid phosphatase and alpha-naphthyl acetate esterase in neoplastic and non-neoplastic lymphocytes. A statistical analysis|url=https://pubmed.ncbi.nlm.nih.gov/6179423|journal=American Journal of Clinical Pathology|volume=78|issue=2|pages=141–149|doi=10.1093/ajcp/78.2.141|issn=0002-9173|pmid=6179423}}</ref> |
| + | * '''Immunophenotype:''' T-cell prolymphocytes exhibit a post-thymic T-cell phenotype. In 60% of cases, the cells are CD4+ and CD8-. In 25% of cases, they co-express both CD4 and CD8, while the remaining 15% are CD4- and CD8+.<ref name=":7" /> | ||
| − | + | Positive (universal) - cyTCL1 (highest specificity), CD2, CD3 (may be weak), CD5, CD7 (strong), TCR-α/β, S100 (30% of cases) | |
| − | + | Positive (subset) - CD4 (in some cases CD4+/CD8+ or CD4-/CD8+), CD52 (usually expressed at high density, therapeutic target), activation markers are variable (CD25, CD38, CD43, CD26, CD27) | |
| − | ( | ||
| − | + | Negative (universal) - TdT, CD1a, CD57, CD16, HTLV1 | |
| − | + | Negative (subset) - CD8 (in some cases CD4+/CD8+ or CD4-/CD8+) | |
| + | ==Links== | ||
| + | (use the "Link" icon that looks like two overlapping circles at the top of the page) <span style="color:#0070C0">(''Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "<nowiki>http://www</nowiki>." portion.'')</span> | ||
| + | ==References== | ||
| + | <references /> | ||
==Notes== | ==Notes== | ||
| − | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. | + | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. |
| − | + | ||
| − | [[Category:HAEM5]][[Category:DISEASE]][[Category:Diseases T]] | + | [[Category:HAEM5]] |
| + | [[Category:DISEASE]] | ||
| + | [[Category:Diseases T]] | ||
Latest revision as of 12:46, 24 March 2025
Haematolymphoid Tumours (WHO Classification, 5th ed.)
Primary Author(s)*
Parastou Tizro, MD, Celeste C. Eno, PhD, Sumire Kitahara, MD
Cedars-Sinai, Los Angeles, CA
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | |
| Category | |
| Family | |
| Type | |
| Subtype(s) |
WHO Essential and Desirable Genetic Diagnostic Criteria
(Instructions: The table will have the diagnostic criteria from the WHO book autocompleted; remove any non-genetics related criteria. If applicable, add text about other classification systems that define this entity and specify how the genetics-related criteria differ.)
| WHO Essential Criteria (Genetics)* | |
| WHO Desirable Criteria (Genetics)* | |
| Other Classification |
*Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the WHO Classification of Tumours.
Related Terminology
(Instructions: The table will have the related terminology from the WHO autocompleted.)
| Acceptable | |
| Not Recommended |
Gene Rearrangements
Put your text here and fill in the table (Instructions: Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Driver Gene | Fusion(s) and Common Partner Genes | Molecular Pathogenesis | Typical Chromosomal Alteration(s) | Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: ABL1 | EXAMPLE: BCR::ABL1 | EXAMPLE: The pathogenic derivative is the der(22) resulting in fusion of 5’ BCR and 3’ABL1. | EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: Common (CML) | EXAMPLE: D, P, T | EXAMPLE: Yes (WHO, NCCN) | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). BCR::ABL1 is generally favorable in CML (add reference). |
| EXAMPLE: CIC | EXAMPLE: CIC::DUX4 | EXAMPLE: Typically, the last exon of CIC is fused to DUX4. The fusion breakpoint in CIC is usually intra-exonic and removes an inhibitory sequence, upregulating PEA3 genes downstream of CIC including ETV1, ETV4, and ETV5. | EXAMPLE: t(4;19)(q25;q13) | EXAMPLE: Common (CIC-rearranged sarcoma) | EXAMPLE: D | EXAMPLE:
DUX4 has many homologous genes; an alternate translocation in a minority of cases is t(10;19), but this is usually indistinguishable from t(4;19) by short-read sequencing (add references). | |
| EXAMPLE: ALK | EXAMPLE: ELM4::ALK
|
EXAMPLE: Fusions result in constitutive activation of the ALK tyrosine kinase. The most common ALK fusion is EML4::ALK, with breakpoints in intron 19 of ALK. At the transcript level, a variable (5’) partner gene is fused to 3’ ALK at exon 20. Rarely, ALK fusions contain exon 19 due to breakpoints in intron 18. | EXAMPLE: N/A | EXAMPLE: Rare (Lung adenocarcinoma) | EXAMPLE: T | EXAMPLE:
Both balanced and unbalanced forms are observed by FISH (add references). | |
| EXAMPLE: ABL1 | EXAMPLE: N/A | EXAMPLE: Intragenic deletion of exons 2–7 in EGFR removes the ligand-binding domain, resulting in a constitutively active tyrosine kinase with downstream activation of multiple oncogenic pathways. | EXAMPLE: N/A | EXAMPLE: Recurrent (IDH-wildtype Glioblastoma) | EXAMPLE: D, P, T | ||
Rearrangements involving the TCL1 (T-cell leukemia/lymphoma 1) family genes—TCL1A, MTCP1 (mature T-cell proliferation), or TCL1B (also known as TCL1/MTCP1-like 1 [TML1])—are highly specific to T-PLL and occur in more than 90% of cases. These translocations juxtapose the TRA locus with the oncogenes TCL1A or TCL1B, or in the case of t(X;14), with the MTCP1 gene.[1][2]
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| inv(14)(q11.2q32.1)
t(14;14)(q11.2;q32.1) |
TCL1A/B ,TRD | inv(14) ~60%
t(14;14) ~25% |
Yes | No | No | These genetic abnormalities serve as diagnostic markers and generally indicate an aggressive disease. This is due to their role in overexpressing oncogenes like TCL1A. Major diagnostic criteria.[1] | |
| t(X;14)(q28;q11.2) | MTCP1, TRD | Low (5%) | Yes | No | No | Major diagnostic criteria.[1] |
Individual Region Genomic Gain/Loss/LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene rearrangements. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Can refer to CGC workgroup tables as linked on the homepage if applicable. Please include references throughout the table. Do not delete the table.)
| Chr # | Gain, Loss, Amp, LOH | Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size] | Relevant Gene(s) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7 |
EXAMPLE:
Unknown |
EXAMPLE: D, P | EXAMPLE: No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add references). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8 |
EXAMPLE:
Unknown |
EXAMPLE: D, P | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add references). | |
| EXAMPLE:
17 |
EXAMPLE: Amp | EXAMPLE:
17q12; chr17:39,700,064-39,728,658 [hg38; 28.6 kb] |
EXAMPLE:
ERBB2 |
EXAMPLE: D, P, T | EXAMPLE:
Amplification of ERBB2 is associated with HER2 overexpression in HER2 positive breast cancer (add references). Add criteria for how amplification is defined. | |
Approximately 70-80% of T-PLL karyotypes are complex, which is considered minor diagnostic criteria, and usually include 3-5 or more structural aberrations. Common cytogenetic abnormalities include those of chromosome 8, such as idic(8)(p11.2), t(8;8)(p11.2;q12), and trisomy 8q. Other frequent changes are deletions in 12p13 and 22q, gains in 8q24 (MYC), and abnormalities in chromosomes 5p, 6, and 17.[3]
Table: A list of clinically significant and/or recurrent CNAs and CN-LOH with potential or strong diagnostic, prognostic and treatment implications in T-PLL are listed below.
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| 8 | Gain | idic(8)(p11)
t(8;8)(p11;q12) trisomy 8q |
idic(8)(p11.2)
t(8;8)(p11.2;q12) trisomy 8q |
Yes | No | No | Recurrent secondary finding (70-80% of cases). Minor diagnostic criteria.[1] |
| 5 | Abnormality | 5p, 5q [4] | Yes | Yes | No | Minor diagnostic criteria.[1] | |
| 6 | Abnormality | gain of 6p, loss of 6q [5][6] | No | No | No | ||
| 11 | Loss | 11q | ch11q21-q23.3 | Yes | Yes | Yes (poor) | Frequent, Minor diagnostic criteria.[1] |
| 12 | Loss | 12p | 12p13 | Yes | Yes | No | Haploinsufficiency of the CDKN1B gene at the 12p13 locus contributes to the development of T-PLL.[7]
Minor diagnostic criteria.[1] |
| 13 | Loss | 13q | 13q14.3 | Yes | No | No | Minor diagnostic criteria.[1] |
| 17 | Loss | 17p | 17p13 | No | Yes | Yes (resistance to therapy) | May include TP53 gene at 17p13.1 [2] |
| 22 | Loss | Monosomy 22
del(22q) |
(most common) |
Yes | No | No | Leading to the dysregulation of genes such as BCL11B, which is crucial in T-cell development and function.[9]
Minor diagnostic criteria.[1] |
Diagnostic criteria
Diagnosis requires either all three major criteria or the first two major criteria along with one minor criterion:[3]
- Major criteria:
- 5 x 109/L cells of T PLL phenotype in peripheral blood or bone marrow
- T cell clonality by molecular or flow cytometry methods
- Abnormalities of 14q32 or Xq28 or expression of TCL1A/B or MTC**
- Minor criteria:
- Abnormalities involving chromosome 11
- Abnormalities in chromosome 8
- Abnormalities in chromosome 5, 12, 13, 22 or complex karyotype
- Involvement of specific sites (spleen, effusions)
**Cases lacking these abnormalities may be referred to as "TCL1 family-negative T-PLL." by some investigators[9]. It is, however, recommended in WHO-HAEM5 that such cases should preferably be classified as peripheral T-cell lymphoma NOS with leukemic involvement (after exclusion of other specific leukemic T-cell entities) as there are insufficient clinicopathological and molecular data to determine the relationship of TCL1 family–negative T-PLL to T-PLL.
Characteristic Chromosomal or Other Global Mutational Patterns
Put your text here and fill in the table (Instructions: Included in this category are alterations such as hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis; microsatellite instability; homologous recombination deficiency; mutational signature pattern; etc. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Chromosomal Pattern | Molecular Pathogenesis | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
EXAMPLE: See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | EXAMPLE: Common (Oligodendroglioma) | EXAMPLE: D, P | ||
| EXAMPLE:
Microsatellite instability - hypermutated |
EXAMPLE: Common (Endometrial carcinoma) | EXAMPLE: P, T | |||
The most common chromosomal abnormality in T-PLL involves an inversion of chromosome 14, with breakpoints at q11.2 and q32.1, observed in about 60-80% of patients and described as inv(14). Additionally, in 10-20% of cases, there is a translocation t(14;14)(q11.2;q32.1).[3] [2]
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| inv(14)(q11q32)
t(14;14)(q11.2;q32.1) |
Yes | No | No | The most common chromosomal abnormality in T-PLL involves an inversion of chromosome 14, with breakpoints at q11.2 and q32.1, observed in about 60-80% of patients and described as inv(14). Additionally, in 10-20% of cases, there is a translocation t(14;14)(q11.2;q32.1) |
Gene Mutations (SNV/INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent or common as well either disease defining and/or clinically significant. If a gene has multiple mechanisms depending on the type or site of the alteration, add multiple entries in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Details on clinical significance such as prognosis and other important information such as concomitant and mutually exclusive mutations can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Gene | Genetic Alteration | Tumor Suppressor Gene, Oncogene, Other | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
| EXAMPLE:EGFR
|
EXAMPLE: Exon 18-21 activating mutations | EXAMPLE: Oncogene | EXAMPLE: Common (lung cancer) | EXAMPLE: T | EXAMPLE: Yes (NCCN) | EXAMPLE: Exons 18, 19, and 21 mutations are targetable for therapy. Exon 20 T790M variants cause resistance to first generation TKI therapy and are targetable by second and third generation TKIs (add references). |
| EXAMPLE: TP53; Variable LOF mutations
|
EXAMPLE: Variable LOF mutations | EXAMPLE: Tumor Supressor Gene | EXAMPLE: Common (breast cancer) | EXAMPLE: P | EXAMPLE: >90% are somatic; rare germline alterations associated with Li-Fraumeni syndrome (add reference). Denotes a poor prognosis in breast cancer. | |
| EXAMPLE: BRAF; Activating mutations | EXAMPLE: Activating mutations | EXAMPLE: Oncogene | EXAMPLE: Common (melanoma) | EXAMPLE: T | ||
Note: A more extensive list of mutations can be found in cBioportal, COSMIC, and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
Although gene mutations beyond TCL1 family alterations are not yet recognized as diagnostic criteria and remain under investigation for T-PLL, the mutational landscape of T-PLL provides valuable insights. These discoveries open up potential avenues for novel targeted therapies in treating this aggressive form of leukemia.
Deletions and mutations of the ATM gene (present in up to 90% of T-PLL cases but typically absent in other mature T-cell malignancies) are considered highly indicative for a diagnosis of suspected TCL1 family-negative T-PLL.[10][11]
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| ATM | TSG | 53% (COSMIC) | ATM mutation/deletion | None specified | Yes | Yes | Yes (PARP inhibitors, NCT03263637) | Since deletions of or missense mutations at the ATM locus are found in up to 80% to 90% of T-PLL cases, ATM alterations can serve as a minor diagnostic criterion.[1][10] |
| FBXW10 | TSG | 72% (COSMIC) | JAK/STAT pathway | None specified | Unknown | Unknown | Unknown | |
| IL2RG, JAK1, JAK3, STAT5B | Oncogene | 8% JAK1
34% JAK3 16% STAT5B 2% IL2RG (COSMIC) (cumulative prevalence of ~ 60%)[12] |
ATM, TP53, Epigenetic modifiers [13][14] | Typically, mutations within this pathway occur in a mutually exclusive manner.[11] | Yes | Yes | Yes | Targeting this pathway with specific JAK/STAT pathway inhibitors, such as tofacitinib, has shown promise in preclinical studies and early clinical trials. Combining JAK/STAT inhibitors with other treatments, like BCL-2 inhibitors, may enhance therapeutic efficacy and improve outcomes for T-PLL patients[15][16] |
| EZH2 | Both oncogene and TSG | 16% (COSMIC) | JAK/STAT pathway[13][14] | None specified | No | No (see note) | See note | EZH2 inhibitors like tazemetostat have shown efficacy in other hematologic malignancies, providing a rationale for their potential use in T-PLL |
| BCOR | TSG | 8% (COSMIC) | JAK/STAT pathway[13][14] | None specified | No | No (see note) | No | A negative impact on overall survival (OS) was not observed for T-PLL patients in the study. However, this might be attributable to the relatively low number of cases compared to studies on AML and MDS.[17] |
| SAMHD1 | TSG | ~7-20%[10][18] | None specified | None specified | Yes | Yes | No | SAMHD1 mutations may indicate a defective DNA damage response and aggressive disease [18] |
| CHEK2 | TSG | 5% (COSMIC) | ATM, TP53, JAK/STAT pathway, Epigenetic modifiers | None specified | No | Yes | No | CHEK2 mutations may indicate a defective DNA damage response and aggressive disease [11][19] |
| TP53 | TSG | 2% (COSMIC) | ATM, JAK/STAT pathway, Epigenetic modifiers | None specified-In a study of T-PLL case, TP53 mutations were predominantly found in patients lacking TCRA/D rearrangements.[17] | No | Yes | Associated with resistance to therapy | Mutations in TP53 are less frequent than deletions.[17]May show overexpression of p53 in some cases.[2] |
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
Epigenomic Alterations
Research indicates that epigenetic modifications in the regulatory regions of key oncogenes and genes involved in DNA damage response and T-cell receptor regulation are clearly present. These changes are closely associated with the transcriptional dysregulation that forms the core lesions of T-PLL.[20]
Genes and Main Pathways Involved
The key pathways involved in the pathogenesis of T-cell prolymphocytic leukemia (T-PLL) include DNA damage repair, T-cell receptor (TCR) signaling, and epigenetic modulation. Additionally, there is frequent mutational activation of the IL2RG-JAK1-JAK3-STAT5B pathway, which plays a significant role in the disease's development and progression.[1]
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| TCL1A/B rearrangement | AKT signaling and TCR signal amplification pathways | Increased cell survival and proliferation |
| MTCP1 | AKT signaling and TCR signal amplification pathways | Increased cell survival and proliferation |
| ATM, CHEK2 | DNA damage repair pathway | Genomic instability |
| JAK1, JAK3, STAT5B | JAK-STAT pathway | Unchecked cell growth and survival |
| IL2RG | JAK-STAT pathway, Cytokine signaling | Promoting lymphocyte proliferation |
| EZH2 | Transcription regulator | Altering the epigenetic landscape |
Genetic Diagnostic Testing Methods
Diagnosing T-PLL involves a combination of clinical evaluation, laboratory tests, imaging studies, and genetic testing to identify diagnostic criteria. T-cell clonality can be confirmed through PCR, NGS, or flow cytometry.[21] Patients with T-PLL often exhibit complex karyotypes with recurrent genetic features that aid in diagnosis. Therefore, cytogenetic studies are useful for distinguishing T-PLL from other T-lymphoproliferative disorders.[1]
- Cytogenetic Analysis
- Karyotyping: To identify characteristic chromosomal abnormalities, such as inv(14)(q11q32), t(14;14)(q11;q32), or other translocations involving chromosome 14. Major diagnostic criteria
- Fluorescence In Situ Hybridization (FISH): To detect specific genetic abnormalities, such as TCL1 gene rearrangements as a Major diagnostic criterion or MYC as a Minor diagnostic criterion (alternatively, molecular studies could be used). see note.
Note: TCL1A break-apart probes specific to the 14q32 region can identify translocations involving TCL1A. When a TCL1A rearrangement is not identified and the patient has T-cell prolymphocytic leukemia/lymphoma (T-PLL), reflex testing using the TRAD break-apart probe set may be performed.
- Molecular Genetic Testing
- Polymerase Chain Reaction (PCR) and Reverse Transcription PCR (RT-PCR): To show the rearrangements of the TR gene (TCRB, TCRG loci) as a Major diagnostic criterion, and alternative to FISH rearrangements of the TCL1 or MTCP genes at the TRD locus can be detected by PCR. Major diagnostic criteria
- Next generation sequencing (NGS)-See note
Note: Although alterations of TCL1A, TCL1B (TML1), or MTCP are present in more than 90% of cases, they are not found in 100% of cases. Taken together, assessment of clonal TCR rearrangement, cytogenetics, and FISH are relevant genetic tests to establish the diagnosis of T-PLL. Genetic sequencing is currently not a diagnostic requirement; however, it may provide information regarding the underlying pathogenesis of T-PLL or might help to identify relevant prognostic subgroups.[1]
Familial Forms
While there is no noticeable familial clustering of T-cell prolymphocytic leukemia (T-PLL), a subset of cases can develop in the context of ataxia-telangiectasia (AT). AT is characterized by germline mutations in the ATM gene, and patients with AT are at an increased risk for various malignancies, including T-PLL. In these cases, biallelic inactivation of the ATM tumor suppressor gene occurs, with about 10% to 15% penetrance of the tumor phenotype by early adulthood. T-PLL represents nearly 3% of all malignancies in patients with ataxia-telangiectasia. [22] [23] [24]
Additional Information
- In T-PLL, the rapid growth of the disease necessitates immediate initiation of treatment. The most effective first-line treatment is alemtuzumab, an anti-CD52 antibody with remission rates over 80%. However, these remissions usually last only 1-2 years. To potentially extend remission, eligible patients are advised to undergo allogeneic blood stem cell transplantation (allo-SCT) during their first complete remission, which can lead to longer remission durations of over 4-5 years for 15-30% of patients. Consequently, the prognosis for T-PLL remains poor, with median overall survival times under two years and five-year survival rates below 5%. Ongoing studies are exploring molecularly targeted drugs and signaling pathway inhibitors, for routine clinical use in treating T-PLL.
This disease is defined/characterized as detailed below:
- T-prolymphocytic leukemia (T-PLL) is an aggressive form of T-cell leukemia marked by the proliferation of small to medium-sized prolymphocytes exhibiting a mature post-thymic T-cell phenotype.[3]
The epidemiology/prevalence of this disease is detailed below:
- T-PLL is an uncommon disease, accounting for approximately 2% of all mature lymphoid leukemias in adults. It mainly affects older individuals, with a median onset age of 65 years, ranging from 30 to 94 years. The disorder exhibits a slight male predominance, with a male to female ratio of 1.33:1.[3]
The clinical features of this disease are detailed below:
- The most prevalent symptom of the disease is a leukemic presentation, characterized by a rapid, exponential increase in lymphocyte counts, which exceed 100 × 10^9/L in 75% of patients. Approximately 30% of patients may initially experience an asymptomatic, slow-progressing phase, but this typically develops into an active disease state.[3][1]
Signs and symptoms - B symptoms (Fever, night sweats, weight loss); Hepatosplenomegaly (Frequently observed); Generalized lymphadenopathy with slightly enlarged lymph nodes (Frequently observed); Cutaneous involvement (20%); Malignant effusions (15%)
Laboratory findings - Anemia and thrombocytopenia; Marked lymphocytosis > 100 × 10^9/L (75% of cases); Atypical lymphocytosis > 5 × 10^9/L; Serum lactate dehydrogenase (LDH) (increased-may reflect disease burden); β2 microglobulin (B2M) (increased-may reflect disease burden)
The sites of involvement of this disease are detailed below:
- Peripheral blood, bone marrow, spleen (mostly red pulp), liver, lymph node (mostly paracortical), and sometimes skin and serosa (primarily pleura). Extra lymphatic and extramedullary atypical manifestations including skin, muscles and intestines are particularly common in relapse.[3]
The morphologic features of this disease are detailed below:
- Blood smears in T-PLL typically reveal anemia, thrombocytopenia, and leukocytosis, with atypical lymphocytes in three morphological forms: The most common form (75% of cases) features medium-sized cells with a high nuclear-to-cytoplasmic ratio, moderately condensed chromatin, a single visible nucleolus, and slightly basophilic cytoplasm. In 20% of cases, the cells appear as a small cell variant with densely condensed chromatin and an inconspicuous nucleolus. About 5% of cases exhibit a cerebriform variant with irregular nuclei resembling those in mycosis fungoides. Regardless of the nuclear features, a common morphological characteristic is the presence of cytoplasmic protrusions or blebs.[25]Bone marrow aspirates show clusters of these neoplastic cells, with a mixed pattern of involvement including diffuse and interstitial, in trephine core biopsy.[1]
The immunophenotype of this disease is detailed below:
- Cytochemistry: T-cell prolymphocytes show strong staining with alpha-naphthyl acetate esterase and acid phosphatase, presenting a distinctive dot-like pattern, but cytochemistry is not commonly used for diagnosis.[26]
- Immunophenotype: T-cell prolymphocytes exhibit a post-thymic T-cell phenotype. In 60% of cases, the cells are CD4+ and CD8-. In 25% of cases, they co-express both CD4 and CD8, while the remaining 15% are CD4- and CD8+.[2]
Positive (universal) - cyTCL1 (highest specificity), CD2, CD3 (may be weak), CD5, CD7 (strong), TCR-α/β, S100 (30% of cases)
Positive (subset) - CD4 (in some cases CD4+/CD8+ or CD4-/CD8+), CD52 (usually expressed at high density, therapeutic target), activation markers are variable (CD25, CD38, CD43, CD26, CD27)
Negative (universal) - TdT, CD1a, CD57, CD16, HTLV1
Negative (subset) - CD8 (in some cases CD4+/CD8+ or CD4-/CD8+)
Links
(use the "Link" icon that looks like two overlapping circles at the top of the page) (Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "http://www." portion.)
References
- ↑ Jump up to: 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Staber, Philipp B.; et al. (2019-10-03). "Consensus criteria for diagnosis, staging, and treatment response assessment of T-cell prolymphocytic leukemia". Blood. 134 (14): 1132–1143. doi:10.1182/blood.2019000402. ISSN 1528-0020. PMC 7042666 Check
|pmc=value (help). PMID 31292114. - ↑ Jump up to: 2.0 2.1 2.2 2.3 2.4 Matutes E, et al., (2017). T-cell prolymphocytic leukemia, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. Revised 4th Edition. IARC Press: Lyon, France, p346-347.
- ↑ Jump up to: 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Elenitoba-Johnson K, et al. T-prolymphocytic leukemia. In: WHO Classification of Tumours Editorial Board. Haematolymphoid tumours [Internet]. Lyon (France): International Agency for Research on Cancer; 2024 [cited 2024 June 12]. (WHO classification of tumors series, 5th ed.; vol. 11). Available from: https://tumourclassification.iarc.who.int/chaptercontent/63/209
- ↑ Tirado, Carlos A.; et al. (2012-08-20). ""T-cell prolymphocytic leukemia (T-PLL), a heterogeneous disease exemplified by two cases and the important role of cytogenetics: a multidisciplinary approach"". Experimental Hematology & Oncology. 1 (1): 21. doi:10.1186/2162-3619-1-21. ISSN 2162-3619. PMC 3514161. PMID 23211026.
- ↑ Dearden, Claire (2012-07-19). "How I treat prolymphocytic leukemia". Blood. 120 (3): 538–551. doi:10.1182/blood-2012-01-380139. ISSN 1528-0020. PMID 22649104.
- ↑ Soulier, J.; et al. (2001-07). "A complex pattern of recurrent chromosomal losses and gains in T-cell prolymphocytic leukemia". Genes, Chromosomes & Cancer. 31 (3): 248–254. doi:10.1002/gcc.1141. ISSN 1045-2257. PMID 11391795. Check date values in:
|date=(help) - ↑ Le Toriellec, Emilie; et al. (2008-02-15). "Haploinsufficiency of CDKN1B contributes to leukemogenesis in T-cell prolymphocytic leukemia". Blood. 111 (4): 2321–2328. doi:10.1182/blood-2007-06-095570. ISSN 0006-4971. PMID 18073348.
- ↑ Stengel, Anna; et al. (2014-12-06). "A Comprehensive Cytogenetic and Molecular Genetic Characterization of Patients with T-PLL Revealed Two Distinct Genetic Subgroups and JAK3 Mutations As an Important Prognostic Marker". Blood. 124 (21): 1639–1639. doi:10.1182/blood.v124.21.1639.1639. ISSN 0006-4971.
- ↑ Jump up to: 9.0 9.1 9.2 Fang, Hong; et al. (2023-09). "T-prolymphocytic leukemia: TCL1 or MTCP1 rearrangement is not mandatory to establish diagnosis". Leukemia. 37 (9): 1919–1921. doi:10.1038/s41375-023-01956-3. ISSN 1476-5551. PMID 37443196 Check
|pmid=value (help). Check date values in:|date=(help) - ↑ Jump up to: 10.0 10.1 10.2 Schrader, A.; et al. (2018-02-15). "Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL". Nature Communications. 9 (1): 697. doi:10.1038/s41467-017-02688-6. ISSN 2041-1723. PMC 5814445. PMID 29449575.
- ↑ Jump up to: 11.0 11.1 11.2 Kiel, Mark J.; et al. (2014-08-28). "Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia". Blood. 124 (9): 1460–1472. doi:10.1182/blood-2014-03-559542. ISSN 1528-0020. PMC 4148768. PMID 24825865.
- ↑ Wahnschaffe, Linus; et al. (2019-11-21). "JAK/STAT-Activating Genomic Alterations Are a Hallmark of T-PLL". Cancers. 11 (12): 1833. doi:10.3390/cancers11121833. ISSN 2072-6694. PMC 6966610. PMID 31766351.
- ↑ Jump up to: 13.0 13.1 13.2 Andersson, E. I.; et al. (2018-03). "Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling". Leukemia. 32 (3): 774–787. doi:10.1038/leu.2017.252. ISSN 1476-5551. PMID 28804127. Check date values in:
|date=(help) - ↑ Jump up to: 14.0 14.1 14.2 Pinter-Brown, Lauren C. (2021-12-30). "JAK/STAT: a pathway through the maze of PTCL?". Blood. 138 (26): 2747–2748. doi:10.1182/blood.2021014238. ISSN 0006-4971.
- ↑ Gomez-Arteaga, Alexandra; et al. (2019-07). "Combined use of tofacitinib (pan-JAK inhibitor) and ruxolitinib (a JAK1/2 inhibitor) for refractory T-cell prolymphocytic leukemia (T-PLL) with a JAK3 mutation". Leukemia & Lymphoma. 60 (7): 1626–1631. doi:10.1080/10428194.2019.1594220. ISSN 1029-2403. PMC 8162842 Check
|pmc=value (help). PMID 30997845. Check date values in:|date=(help) - ↑ . doi:10.1182/blood.v126.23.5486.5486 https://ashpublications.org/blood/article/126/23/5486/134544/Refractory-TCell-Prolymphocytic-Leukemia-with-JAK3. Missing or empty
|title=(help) - ↑ Jump up to: 17.0 17.1 17.2 Stengel, Anna; et al. (2016-01). "Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker". Genes, Chromosomes & Cancer. 55 (1): 82–94. doi:10.1002/gcc.22313. ISSN 1098-2264. PMID 26493028. Check date values in:
|date=(help) - ↑ Jump up to: 18.0 18.1 Johansson, Patricia; et al. (2018-01-19). "SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia". Blood Cancer Journal. 8 (1): 11. doi:10.1038/s41408-017-0036-5. ISSN 2044-5385. PMC 5802577. PMID 29352181.
- ↑ Braun, Till; et al. (2021). "Advanced Pathogenetic Concepts in T-Cell Prolymphocytic Leukemia and Their Translational Impact". Frontiers in Oncology. 11: 775363. doi:10.3389/fonc.2021.775363. ISSN 2234-943X. PMC 8639578 Check
|pmc=value (help). PMID 34869023 Check|pmid=value (help). - ↑ Tian, Shulan; et al. (2021-04-15). "Epigenetic alteration contributes to the transcriptional reprogramming in T-cell prolymphocytic leukemia". Scientific Reports. 11 (1): 8318. doi:10.1038/s41598-021-87890-9. ISSN 2045-2322. PMC 8050249 Check
|pmc=value (help). PMID 33859327 Check|pmid=value (help). - ↑ Kotrova, Michaela; et al. (2018-11). "Next-generation amplicon TRB locus sequencing can overcome limitations of flow-cytometric Vβ expression analysis and confirms clonality in all T-cell prolymphocytic leukemia cases". Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 93 (11): 1118–1124. doi:10.1002/cyto.a.23604. ISSN 1552-4930. PMID 30414304. Check date values in:
|date=(help) - ↑ Suarez, Felipe; et al. (2015-01-10). "Incidence, presentation, and prognosis of malignancies in ataxia-telangiectasia: a report from the French national registry of primary immune deficiencies". Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 33 (2): 202–208. doi:10.1200/JCO.2014.56.5101. ISSN 1527-7755. PMID 25488969.
- ↑ Taylor, A. M.; et al. (1996-01-15). "Leukemia and lymphoma in ataxia telangiectasia". Blood. 87 (2): 423–438. ISSN 0006-4971. PMID 8555463.
- ↑ Li, Geling; et al. (2017-12-26). "T-cell prolymphocytic leukemia in an adolescent with ataxia-telangiectasia: novel approach with a JAK3 inhibitor (tofacitinib)". Blood Advances. 1 (27): 2724–2728. doi:10.1182/bloodadvances.2017010470. ISSN 2473-9529. PMC 5745136. PMID 29296924.
- ↑ Gutierrez, Marc; et al. (2023-07-28). "T-Cell Prolymphocytic Leukemia: Diagnosis, Pathogenesis, and Treatment". International Journal of Molecular Sciences. 24 (15): 12106. doi:10.3390/ijms241512106. ISSN 1422-0067. PMC PMC10419310 Check
|pmc=value (help). PMID 37569479 Check|pmid=value (help).CS1 maint: PMC format (link) - ↑ Yang, K.; et al. (1982-08). "Acid phosphatase and alpha-naphthyl acetate esterase in neoplastic and non-neoplastic lymphocytes. A statistical analysis". American Journal of Clinical Pathology. 78 (2): 141–149. doi:10.1093/ajcp/78.2.141. ISSN 0002-9173. PMID 6179423. Check date values in:
|date=(help)
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.