Difference between revisions of "HAEM5:B-lymphoblastic leukaemia/lymphoma with hypodiploidy"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:B-lymphoblastic leukaemia/lymphoma with hypodiploidy}} | {{DISPLAYTITLE:B-lymphoblastic leukaemia/lymphoma with hypodiploidy}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
{{Under Construction}} | {{Under Construction}} | ||
| − | <blockquote class='blockedit'>{{Box-round|title= | + | <blockquote class='blockedit'>{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:B-Lymphoblastic Leukemia/Lymphoma with Hypodiploidy]]. |
}}</blockquote> | }}</blockquote> | ||
| + | |||
| + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | ||
| + | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| Line 16: | Line 19: | ||
__TOC__ | __TOC__ | ||
| − | == | + | ==WHO Classification of Disease== |
| − | B | + | {| class="wikitable" |
| − | + | !Structure | |
| − | + | !Disease | |
| − | + | |- | |
| − | + | |Book | |
| + | |Haematolymphoid Tumours (5th ed.) | ||
| + | |- | ||
| + | |Category | ||
| + | |B-cell lymphoid proliferations and lymphomas | ||
| + | |- | ||
| + | |Family | ||
| + | |Precursor B-cell neoplasms | ||
| + | |- | ||
| + | |Type | ||
| + | |B-lymphoblastic leukaemias/lymphomas | ||
| + | |- | ||
| + | |Subtype(s) | ||
| + | |B-lymphoblastic leukaemia/lymphoma with hypodiploidy | ||
| + | |} | ||
==Definition / Description of Disease== | ==Definition / Description of Disease== | ||
| Line 42: | Line 59: | ||
==Clinical Features== | ==Clinical Features== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |EXAMPLE Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | EXAMPLE B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | EXAMPLE Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | EXAMPLE Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |EXAMPLE Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | EXAMPLE Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 85: | Line 102: | ||
|Positive||CD10 | |Positive||CD10 | ||
|- | |- | ||
| − | |Negative (universal)||EXAMPLE CD3 | + | |Negative (universal)||<span class="blue-text">EXAMPLE:</span> CD3 |
|- | |- | ||
| − | |Negative (subset)||EXAMPLE CD4 | + | |Negative (subset)||<span class="blue-text">EXAMPLE:</span> CD4 |
|} | |} | ||
| Line 102: | Line 119: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 135: | Line 152: | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain / Loss / LOH== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 145: | Line 162: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |EXAMPLE Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 158: | Line 175: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |EXAMPLE Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 175: | Line 192: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 194: | Line 211: | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| − | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis'')</span> | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 204: | Line 221: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 210: | Line 227: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 248: | Line 265: | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV / INDEL)== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 258: | Line 275: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE: TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | EXAMPLE: | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | EXAMPLE: BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |EXAMPLE: TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |EXAMPLE: 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE: 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |EXAMPLE: IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |EXAMPLE: EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 306: | Line 323: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table.'')</span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |EXAMPLE: BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |EXAMPLE: MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |EXAMPLE: Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |EXAMPLE: CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |EXAMPLE: Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |EXAMPLE: Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |EXAMPLE: KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |EXAMPLE: Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |EXAMPLE: Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
Latest revision as of 17:25, 6 September 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:B-Lymphoblastic Leukemia/Lymphoma with Hypodiploidy.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Ashwini Yenamandra PhD FACMG
Lisa Smith PhD FACMG
Yassmine Akkari PhD FACMG
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | B-cell lymphoid proliferations and lymphomas |
| Family | Precursor B-cell neoplasms |
| Type | B-lymphoblastic leukaemias/lymphomas |
| Subtype(s) | B-lymphoblastic leukaemia/lymphoma with hypodiploidy |
Definition / Description of Disease
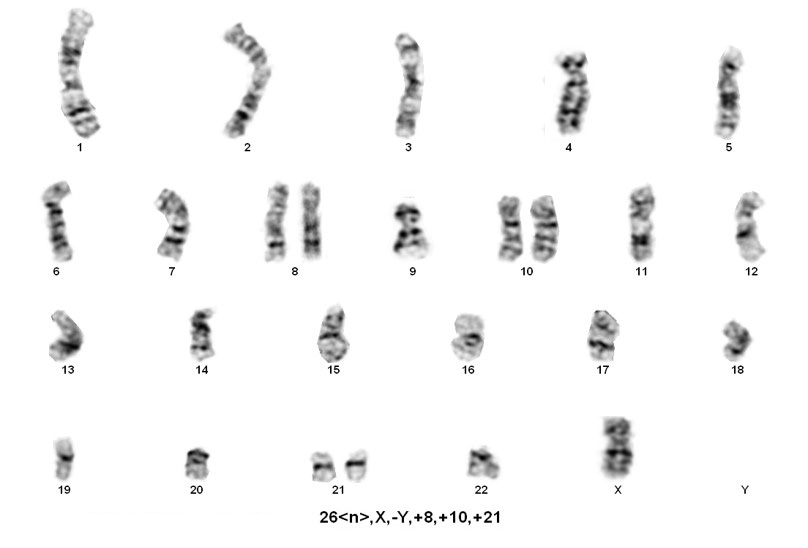
Hypodiploidy is a rare entity comprising approximately 5% of all B-cell acute lymphoblastic leukemias. The majority of cases (>80%) fall within the 44-45 chromosome range. However, beyond this, there are three main groups, that although rare, are associated with a very poor clinical prognosis.
Synonyms / Terminology
Hypodiploidy- High Hypodiploidy, Low Hypodiploidy, Near-haploidy
Epidemiology / Prevalence
Hypodiploid ALL accounts for about 5% of ALL cases. Approximately 1% of ALL cases with hypodiploidy have <45 chromosomes[1]. It is more common in children than adults and accounts for 75% of pediatric cancers[2]. Hypodiploid ALL with chromosome numbers less than 44 is associated with poor prognosis.
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.The clinical features are generally similar to those seen in other types of B-ALL like anemia, thrombocytopenia and neutropenia. May have very high WBC count at presentation. Malignant and poorly differentiated lymphoid cells in the bone marrow, peripheral blood and extramedullary sites[3]. Symptoms may include anemia, thrombocytopenia, leukopenia, fever, weight loss, night sweats, bleeding, bruising, fatigue[3]. Splenomegaly and hepatomegaly (20%), central nervous system (CNS) in 5-8% of patients[3]. Diagnosis: 20% of blasts or more lymphoblasts in the bone marrow, or peripheral blood[3].
Sites of Involvement
Bone Marrow, Peripheral Blood, extra medullary sites.
Morphologic Features
Malignant and poorly differentiated lymphoid cells in the bone marrow, peripheral blood and extramedullary sites[3]. Symptoms may include anemia, thrombocytopenia, leukopenia, fever, weight loss, night sweats, bleeding, bruising, fatigue[3]. Splenomegaly and hepatomegaly (20%), central nervous system (CNS) in 5-8% of patients[3]. Diagnosis: 20% of blasts or more lymphoblasts in the bone marrow, or peripheral blood[3]
Immunophenotype
Put your text here and/or fill in the table
| Finding | Marker |
|---|---|
| Positive | CD19 |
| Positive | CD10 |
| Negative (universal) | EXAMPLE: CD3 |
| Negative (subset) | EXAMPLE: CD4 |
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.N/A
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
B-lymphoblastic leukemia/lymphoma (B-ALL/LBL) is the most common cause of cancer in pediatric patients. It is characterized by recurrent genetic abnormalities of chromosome number, deletions, duplications and translocations. Hypodiploidy, a neoplasm of lymphoblasts containing less than 46 chromosomes[1]. Hypodiploid ALL has poor prognosis and near haploid with worst prognosis[1][4][5].
Patients with 44 chromosomes had a better event free survival (EFS) than patients with fewer than 44 chromosomes[5]. However, patients with 44 chromosomes and monosomy 7 or a dicentric chromosome had worse EFS[5]. Children and adults with less than 44 chromosomes had poor outcome despite contemporary therapy[5].
In near haploid cases, two-thirds had activation of RAS signaling and P13K signaling pathways; these are sensitive to P13K inhibitors indicating these drugs may offer a new therapeutic option[4].
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.
Chromosome Number Gain/Loss/Amp/LOH Region 17 Gain SOX9 9 Loss CDKN2A/CDKN2B (22-50%)
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.Based on WHO classification[1], hypodiploidy is divided into:
1. Near-haploidy (NH=23-29 chromosomes) This is a very rare category, has been observed in the pediatric population with virtually no adult cases reported. Nonrandom retention of the X chromosome plus chromosomes 8, 14, 18, and 21 are frequently observed.
2. Low Hypodiploidy (LH=33-39 chromosomes) This category was reported in both children and adults. Nonrandom retention of two copies of chromosomes from the following: the sex chromosomes plus chromosomes 1,6, 8, 10, 14, 18, and19. Chromosome 21 is almost always retained in two copies.
3. High Hypodiploidy (HH=40-43 chromosomes) This category was observed in both children and adults. Chromosome abnormalities include whole chromosome loss, specifically one sex chromosome and often chromosomes 7, 9, and/or 13. Also detected are structural anomalies especially dicentric chromosomes involving chromosomes 7, 9 or 12.
4. Near-diploid (ND=44-45 chromosomes) This category is not often included in hypodiploid.
Note: A slight variation in the range of chromosome number has been reported in the literature in the classification of NH, LH, HH and NH[1][3][4][6][5][7][8][9][10][11][12] [13][14][15][16][17][2] [1-17].
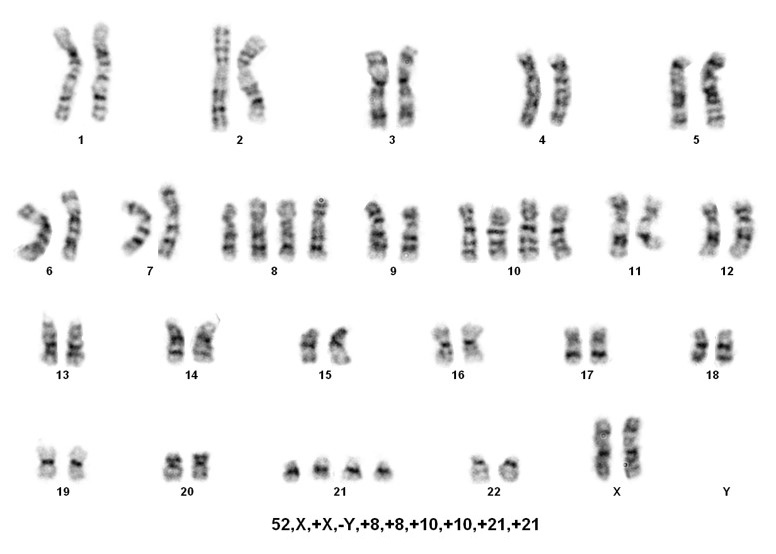
Sorting patients into these three rare groups is easy. However, detecting the presence of a masked low-hypodiploid/masked near-hypodiploid group, which is endoreduplication of the low- and near-haploid groups and associated with a very poor prognosis, is difficult. Often karyotypes in these two groups, usually ranging from 56-78 chromosomes, are mistaken for hyperdiploidy/near-triploidy, which in itself is associated with a good prognosis. The key is to look for trisomies vs tetrasomies of the chromosomes. Typically, hyperdiploidy/near-triploidy should have three copies of several chromosomes (usually the X, 4, 10, 17, and 18), and four copies of 14 and 21. However, the masked low-hypodiploid/masked near-hypodiploid groups should show tetrasomies for the sex chromosomes and chromosomes 1, 14, 18, 21, and 22 while having only two copies of chromosomes 7 and 17.
When only a 56-78 chromosome count is detected, the above mentioned criteria is helpful, but SNP-array testing can also be informative. Masked near-haploidy appears to show LOH involving the chromosomes that are not gained and true hyperdiploidy will show heterozygosity. Dicentric chromosomes reportedly originated from chromosomes 9p, 12p or 20q in near diploid karyotypes[4]. In near haploid aneuploidy of chromosomes 1 through 7, 9, 11, 13, 15-17, 19,20, 22 while in low hypodiploid aneuploidy of chromosomes 2 through 4, 7, 9, 12, 13, 15 and 17 were reported[4].
Near haploidy may be the primary event with loss of chromosomes, followed by a secondary event of doubling of chromosomes indicating uniparental isodisomy (UPID), microdeletions if any may occur after the secondary event[6].
In hypodiploid ALL, molecular mutations are equally as important as chromosome number, or as a result of chromosome number, molecular mutations have a driving effect. Both LH and NH have common mutations involved in the disease process. In near haploid ALL (NH) RTK and RAS (71%) signaling were a hallmark[4]. In addition, lymphoid transcription factor gene IKZF3 (13%, encoding AIOLOS) and deletions of histone cluster at 6p22 (19%) were also reported[4]. In low hypodiploid (LH) ALL mutations involved TP53 (91.2%) and IKZF2 (53%, encoding HELIOS, 2q34), and RB1 genes (41%) loci[4]. Both NH and LH had activation of RAS signaling and P13K signaling pathways and sensitive to P13K inhibitors indicating these drugs may offer a new therapeutic option[4]. Inn this group, several studies have not only identified a high percentage of pediatric patients with TP53 mutations, but close to half displayed germline mutations, suggesting that LH ALL is a manifestation of Li-Fraumeni syndrome in children. Adults also showed a high incidence of TP53 (91%) in low hypodiploid ALL mutations, but these mutations appear to be somatic in origin. In NH, mutations appear in genes involving receptor tyrosine kinase (RTK) pathway, Ras signaling, IKZF3 (17q21.1), and histone clusters, but rarely IZFK2, RB1, or TP53[4].
Copy number alterations and sequence mutations have been reported in FLT3, NF1, KRAS, NRAS, PTPN11, RTK, RAS, IKZF1, IKZF2, IKZF3, TP53, RB1, Histone, 6q22, CDKN2A, CDKN2B, PAX5, and PAG1 gene loci[4].
The most significant observation by Holmfeldt et al.,[4] is that a global difference in the gene expression profiles distinguishes subgroups of hypodiploid ALL. More than 600 genes had subtype specific enrichment on gene set enrichment analysis[4]. In addition, RAS pathway, RB1 and TP53 mutations mimic solid tumor pathways[4].
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.
Gene Mutation Oncogene/Tumor Suppressor/Other Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) Prevalence (COSMIC/TCGA/Other) TP53 R280S, Y220C and several other mutations, please see reference[4]. Tumor Suppressor Missense/Nonsense/Insertion/Splice in Low Hypodiploid about 90% Other Mutations
RTK-RAS signaling pathways: About two-thirds of near haploid ALL (71%) had activation of RTK-RAS signaling pathways including deletion, amplification and sequence mutation of NF1, NRAS, KRAS, MAPK1, FLT3 and PTPN11[4]. NF1 mutation was reported in 44% of near haploid cases with a biallelic mutation of NF1 in 77% of the near haploid cases. In 68% of the cases, the NF1 deletions were intragenic involving exons 15 through 35[4]. The focal deletion results in deletion of GAP[4].
PAG1 mutations: Recurrent alterations of PAG1 was reported in 10.3% of near haploid ALL, PAG1 mutations are rare in other hypodiploid cases[4]. PAG1 was identified as a putative RAS signaling inhibitor and have a negative regulatory function in proximal B-cell receptor signaling[4].
TP53 mutations: High mutation rate was observed (91%) in low hypodiploid than in non-low hypodiploid (5%) B-ALL; In low hypodiploid ALL, 43% were observed in non-tumor hematopoietic cells, suggesting either an inherited or a germline de novo origin of the mutation[4].
Epigenomic Alterations
In near haploid 19% of the cases had focal deletions of histone gene cluster at 6p22, however, non-hypodiploid ALL had 8%, lower frequency of these deletions[4].
Of the 25 next generation sequenced haploid cases 16 (64%) cases had twenty six histone modifier gene mutations and of the 15 low hypodiploid ALL cases 9 (60%) cases had 9 mutations; the most common mutation (32%) of the near haploid cases was transcriptional co-activator and histone acetyltransferase CREBBP[4].
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.RTK and RAS pathway alterations as de novo germline mutations or in primitive hematopoietic progenitor cells[4].
Genetic Diagnostic Testing Methods
FLOW, Hematopathology, Cytogenetics, Fluorescence insitu Hybridization (FISH), Next Generation sequencing (NGS), Exome sequencing, Microarray.
Familial Forms
In Low hypodiploid (LH), several studies have not only identified a high percentage of pediatric patients with TP53 mutations, but close to half displayed germline mutations, suggesting that LH ALL is a manifestation of Li-Fraumeni syndrome in children.
Adults also showed a high incidence of TP53 mutations, but these mutations appear to be somatic in origin. In NH, mutations of genes of receptor tyrosine kinase (RTK) pathway, Ras signaling, IKZF3 (17q21.1) and histone clusters, but mutations of IZFK2, RB1, or TP53 were rare.
Additional Information
Genetic abnormalities involving TP53, RB1 and IKZF2 are hallmarks of low hypodiploid ALL, where as near haploid ALL has RTK, RAS and IKZF3 alterations[4].
Links
N/A
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 1.2 1.3 1.4 Borowitz MJ, et al., (2017). B-Lymphoblastic leukaemia/lymphoma with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p206.
- ↑ 2.0 2.1 Karen Seiter, MD; Chief Editor: Emmanuel C Besa, MD (2018). Acute lymphoblastic leukemia (ALL). Medscape. emedicine, Medscape Article, Drugs & Diseases, Hematology.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Terwilliger, T.; et al. (2017). "Acute lymphoblastic leukemia: a comprehensive review and 2017 update". Blood Cancer Journal. 7 (6): e577. doi:10.1038/bcj.2017.53. ISSN 2044-5385. PMC 5520400. PMID 28665419.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 4.20 4.21 4.22 4.23 4.24 Holmfeldt, Linda; et al. (2013). "The genomic landscape of hypodiploid acute lymphoblastic leukemia". Nature Genetics. 45 (3): 242–252. doi:10.1038/ng.2532. ISSN 1546-1718. PMC 3919793. PMID 23334668.
- ↑ 5.0 5.1 5.2 5.3 5.4 Nachman, James B.; et al. (2007). "Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia". Blood. 110 (4): 1112–1115. doi:10.1182/blood-2006-07-038299. ISSN 0006-4971. PMC 1939895. PMID 17473063.
- ↑ 6.0 6.1 Safavi, S.; et al. (2013). "Loss of chromosomes is the primary event in near-haploid and low-hypodiploid acute lymphoblastic leukemia". Leukemia. 27 (1): 248–250. doi:10.1038/leu.2012.227. ISSN 1476-5551. PMID 22889820.
- ↑ Safavi, Setareh; et al. (2017). "Near-haploid and low-hypodiploid acute lymphoblastic leukemia: two distinct subtypes with consistently poor prognosis". Blood. 129 (4): 420–423. doi:10.1182/blood-2016-10-743765. ISSN 1528-0020. PMID 27903530.
- ↑ Mehta, Parinda A.; et al. (2015). "Transplantation Outcomes for Children with Hypodiploid Acute Lymphoblastic Leukemia". Biology of Blood and Marrow Transplantation: Journal of the American Society for Blood and Marrow Transplantation. 21 (7): 1273–1277. doi:10.1016/j.bbmt.2015.04.008. ISSN 1523-6536. PMC 4465998. PMID 25865650.
- ↑ Mullighan, Charles G. (2012). "Molecular genetics of B-precursor acute lymphoblastic leukemia". The Journal of Clinical Investigation. 122 (10): 3407–3415. doi:10.1172/JCI61203. ISSN 1558-8238. PMC 3461902. PMID 23023711.
- ↑ Harrison, Christine J.; et al. (2004). "Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia". British Journal of Haematology. 125 (5): 552–559. doi:10.1111/j.1365-2141.2004.04948.x. ISSN 0007-1048. PMID 15147369.
- ↑ Wang, Yunhong; et al. (2016). "Genome-Wide Single-Nucleotide Polymorphism Array Analysis Improves Prognostication of Acute Lymphoblastic Leukemia/Lymphoma". The Journal of molecular diagnostics: JMD. 18 (4): 595–603. doi:10.1016/j.jmoldx.2016.03.004. ISSN 1943-7811. PMID 27161658.
- ↑ Safavi, Setareh; et al. (2015). "Genetic and epigenetic characterization of hypodiploid acute lymphoblastic leukemia". Oncotarget. 6 (40): 42793–42802. doi:10.18632/oncotarget.6000. ISSN 1949-2553. PMC 4767471. PMID 26544893.
- ↑ Moorman, Anthony V. (2016). "New and emerging prognostic and predictive genetic biomarkers in B-cell precursor acute lymphoblastic leukemia". Haematologica. 101 (4): 407–416. doi:10.3324/haematol.2015.141101. ISSN 1592-8721. PMC 5004393. PMID 27033238.
- ↑ Fang, Min; et al. (2015). "Adult Low-Hypodiploid Acute B-Lymphoblastic Leukemia With IKZF3 Deletion and TP53 Mutation: Comparison With Pediatric Patients". American Journal of Clinical Pathology. 144 (2): 263–270. doi:10.1309/AJCPW83OXPYKPEEN. ISSN 1943-7722. PMID 26185311.
- ↑ Mühlbacher, Verena; et al. (2014). "Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%". Genes, Chromosomes & Cancer. 53 (6): 524–536. doi:10.1002/gcc.22163. ISSN 1098-2264. PMID 24619868.
- ↑ Woo, Jennifer S.; et al. (2014). "Childhood B-acute lymphoblastic leukemia: a genetic update". Experimental Hematology & Oncology. 3: 16. doi:10.1186/2162-3619-3-16. ISSN 2162-3619. PMC 4063430. PMID 24949228.
- ↑ Collins-Underwood, J. R.; et al. (2010). "Genomic profiling of high-risk acute lymphoblastic leukemia". Leukemia. 24 (10): 1676–1685. doi:10.1038/leu.2010.177. ISSN 1476-5551. PMID 20739952.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.
[[Copy Number and cn-LOH Abnormalities in ALL]
*Citation of this Page: “B-lymphoblastic leukaemia/lymphoma with hypodiploidy”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 09/6/2024, https://ccga.io/index.php/HAEM5:B-lymphoblastic_leukaemia/lymphoma_with_hypodiploidy.