Difference between revisions of "HAEM5:Chronic myelomonocytic leukaemia"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Chronic myelomonocytic leukaemia}} | {{DISPLAYTITLE:Chronic myelomonocytic leukaemia}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
{{Under Construction}} | {{Under Construction}} | ||
| − | <blockquote class='blockedit'>{{Box-round|title= | + | <blockquote class='blockedit'>{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Chronic Myelomonocytic Leukemia (CMML)]]. |
}}</blockquote> | }}</blockquote> | ||
| + | |||
| + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | ||
| + | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| Line 12: | Line 15: | ||
__TOC__ | __TOC__ | ||
| − | == | + | ==WHO Classification of Disease== |
| − | + | {| class="wikitable" | |
| − | + | !Structure | |
| − | + | !Disease | |
| − | + | |- | |
| − | + | |Book | |
| + | |Haematolymphoid Tumours (5th ed.) | ||
| + | |- | ||
| + | |Category | ||
| + | |Myeloid proliferations and neoplasms | ||
| + | |- | ||
| + | |Family | ||
| + | |Myelodysplastic/myeloproliferative neoplasms | ||
| + | |- | ||
| + | |Type | ||
| + | |N/A | ||
| + | |- | ||
| + | |Subtype(s) | ||
| + | |Chronic myelomonocytic leukaemia | ||
| + | |} | ||
==Definition / Description of Disease== | ==Definition / Description of Disease== | ||
| Line 44: | Line 61: | ||
==Clinical Features== | ==Clinical Features== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |EXAMPLE Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | EXAMPLE B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | EXAMPLE Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | EXAMPLE Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |EXAMPLE Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | EXAMPLE Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 64: | Line 81: | ||
<blockquote class='blockedit'>{{Box-round|title=v4:Clinical Features|The content below was from the old template. Please incorporate above.}} | <blockquote class='blockedit'>{{Box-round|title=v4:Clinical Features|The content below was from the old template. Please incorporate above.}} | ||
| − | The clinical features are variable and significantly related to blood cell counts<ref name=":1" /><ref name=":2">Arber DA, et al., (2017). Introduction and overview of the classification of myeloid neoplasms, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Editors. IARC Press: Lyon, France, p82-86.</ref>. Some clinical presentations may be different in dysplastic (WBC count < 13 x 10<sup>9</sup>/L) and proliferative (WBC count ≥ 13 x 10<sup>9</sup>/L) subgroups<ref name=":0" />. In patients with increased white blood cell (WBC) count (~50% of all cases), constitutional symptoms (e.g. weight loss, fever, and night sweats) are common. In cases with cytopenia(s), similar to [[Myelodysplastic Syndromes (MDS)|MDS]], clinical presentations are usually related to insufficient blood cells (anemia, leukopenia and/or thrombocytopenia, e.g. fatigue, infection, and bleeding tendency)<ref>Storniolo AM, Moloney WC, Rosenthal DS, Cox C, Bennett JM. Chronic myelomonocytic leukemia. ''Leukemia''. 1990;4(11):766‐770.</ref>. Hepatosplenomegaly can be present in both subgroups, but are frequently related to leukocytosis. Rare cases with life-threatening hyperleukocytosis have been reported. | + | The clinical features are variable and significantly related to blood cell counts<ref name=":1" /><ref name=":2">Arber DA, et al., (2017). Introduction and overview of the classification of myeloid neoplasms, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Editors. IARC Press: Lyon, France, p82-86.</ref>. Some clinical presentations may be different in dysplastic (WBC count < 13 x 10<sup>9</sup>/L) and proliferative (WBC count ≥ 13 x 10<sup>9</sup>/L) subgroups<ref name=":0" />. In patients with increased white blood cell (WBC) count (~50% of all cases), constitutional symptoms (e.g. weight loss, fever, and night sweats) are common. In cases with cytopenia(s), similar to [[HAEM4:Myelodysplastic Syndromes (MDS)|MDS]], clinical presentations are usually related to insufficient blood cells (anemia, leukopenia and/or thrombocytopenia, e.g. fatigue, infection, and bleeding tendency)<ref>Storniolo AM, Moloney WC, Rosenthal DS, Cox C, Bennett JM. Chronic myelomonocytic leukemia. ''Leukemia''. 1990;4(11):766‐770.</ref>. Hepatosplenomegaly can be present in both subgroups, but are frequently related to leukocytosis. Rare cases with life-threatening hyperleukocytosis have been reported. |
</blockquote> | </blockquote> | ||
| Line 144: | Line 161: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 192: | Line 209: | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain / Loss / LOH== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 202: | Line 219: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |EXAMPLE Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 215: | Line 232: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |EXAMPLE Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 232: | Line 249: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 262: | Line 279: | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| − | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis'')</span> | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 272: | Line 289: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 278: | Line 295: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 290: | Line 307: | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV / INDEL)== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 300: | Line 317: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE: TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | EXAMPLE: | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | EXAMPLE: BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |EXAMPLE: TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |EXAMPLE: 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE: 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |EXAMPLE: IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |EXAMPLE: EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 386: | Line 403: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table.'')</span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |EXAMPLE: BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |EXAMPLE: MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |EXAMPLE: Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |EXAMPLE: CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |EXAMPLE: Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |EXAMPLE: Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |EXAMPLE: KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |EXAMPLE: Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |EXAMPLE: Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
==Genetic Diagnostic Testing Methods== | ==Genetic Diagnostic Testing Methods== | ||
| Line 422: | Line 439: | ||
==Links== | ==Links== | ||
| − | Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page <span style="color:#0070C0">(''Instructions: | + | Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page <span style="color:#0070C0">(''Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "<nowiki>http://www</nowiki>." portion.'')</span> |
==References== | ==References== | ||
Latest revision as of 17:17, 6 September 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Chronic Myelomonocytic Leukemia (CMML).
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Linsheng Zhang, MD, PhD
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Myelodysplastic/myeloproliferative neoplasms |
| Type | N/A |
| Subtype(s) | Chronic myelomonocytic leukaemia |
Definition / Description of Disease
Chronic myelomonocytic leukemia (CMML) is a myelodysplastic/myeloproliferative neoplasm (MDS/MPN) with persistent monocytosis (absolute monocyte count ≥1 x 109/L and ≥10% of leukocytes are monocytes). When myelodysplasia is absent or minimal, clonal evidence(s) of acquired cytogenetic or molecular genetic change(s) in hematopoietic cells, or unexplained monocytosis persisted for ≥3 months supports the diagnosis of CMML.
Before a diagnosis of CMML can be rendered, other well-defined myeloid neoplasms that sometimes present with monocytosis, such as BCR-ABL1-positive chronic myeloid leukemia (especially with p190 fusion) and myeloid/lymphoid neoplasms with rearrangement of PDGFRA, PDGFRB, FGFR1 or PCM1-JAK2 must be ruled out. Dysplastic features are usually seen in at least one myeloid lineage. By definition, the blast percentage must be <20%.
CMML can be subdivided into three categories based on the percentage of blasts and promonocytes (considered blast equivalent) in the peripheral blood and bone marrow[1]:
CMML-0: < 2% blasts in the blood and < 5% in the bone marrow; and no Auer rods.
CMML-1: 2-4% blasts in the blood or 5-9% in the bone marrow; and no Auer rods.
CMML-2: 5-19% blasts in the blood, 10-19% in the bone marrow or Auer rods are present.
Synonyms / Terminology
Put your text here (Instructions: Include currently used terms and major historical ones, adding “(historical)” after the latter.)
Epidemiology / Prevalence
Historically CMML was classified as a subtype of MDS. The exact incidence of CMML is difficult to calculate based on the published data. Recent studies reported approximate incidence of 0.4 cases per 100,000 population[2]. The median patient age at diagnosis is 65-75 years, with a slight male dominance (male-to female ratio of 1.5-3:1)[3][4][5].
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.The clinical features are variable and significantly related to blood cell counts[3][1]. Some clinical presentations may be different in dysplastic (WBC count < 13 x 109/L) and proliferative (WBC count ≥ 13 x 109/L) subgroups[4]. In patients with increased white blood cell (WBC) count (~50% of all cases), constitutional symptoms (e.g. weight loss, fever, and night sweats) are common. In cases with cytopenia(s), similar to MDS, clinical presentations are usually related to insufficient blood cells (anemia, leukopenia and/or thrombocytopenia, e.g. fatigue, infection, and bleeding tendency)[6]. Hepatosplenomegaly can be present in both subgroups, but are frequently related to leukocytosis. Rare cases with life-threatening hyperleukocytosis have been reported.
Sites of Involvement
Blood and bone marrow (always involved).
The common sites of extramedullary infiltrate include spleen, liver, skin and lymph nodes[3][1][7], involvement of other organs, although less common, have also been reported.
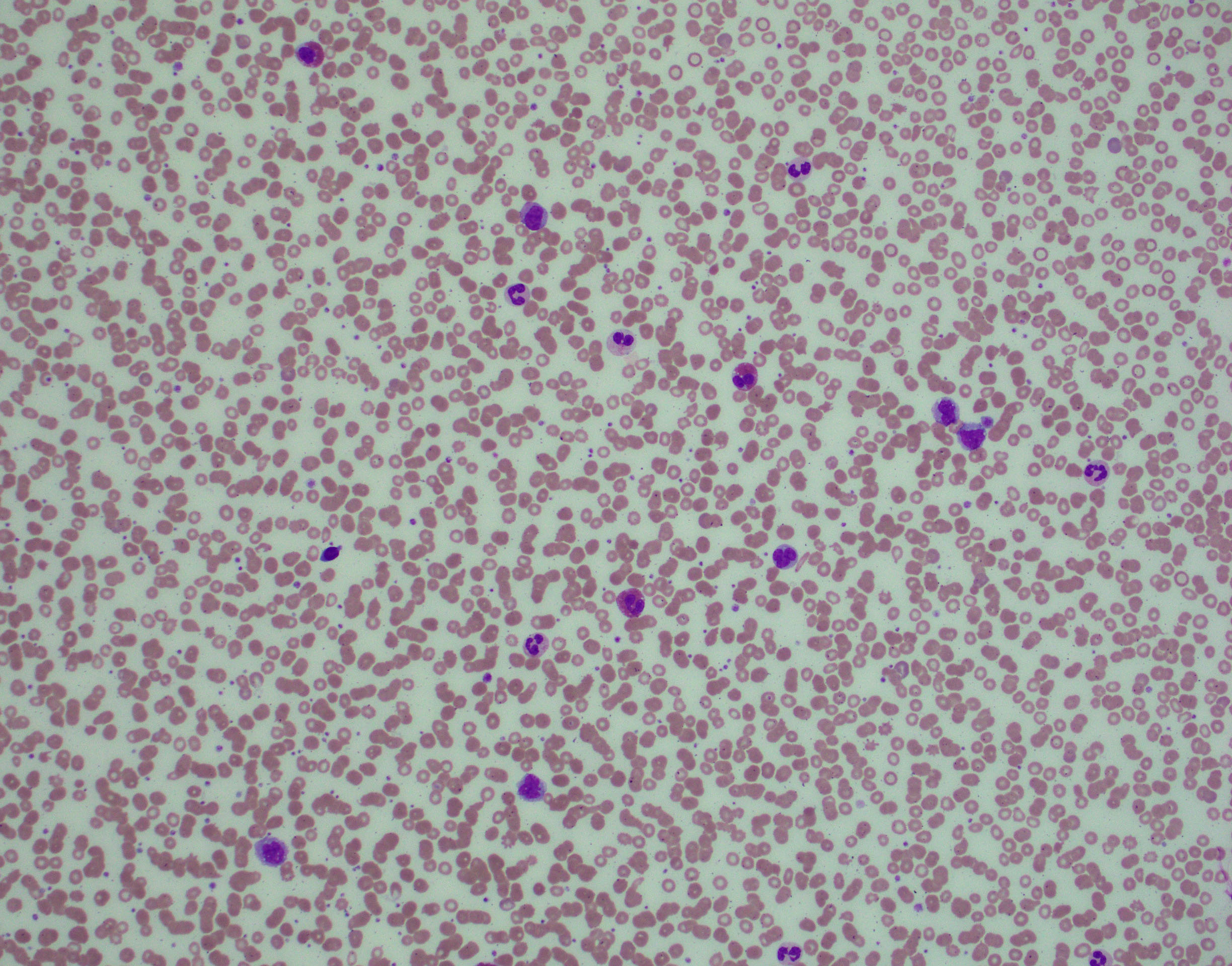
Morphologic Features
Blood:
- Monocytosis ≥ 1 x 109/L and monocytes account for ≥ 10% of the leukocytes.
Monocyte morphology ranges from normal to atypical: unusual nuclear segmentation or chromatin patterns, and abnormal cytoplasmic granulation.
- Blasts and promonocytes together must be <20% of the leukocytes.
- Variable changes in other blood cells are usually present.
- WBC count: increased, normal to slightly decreased, with neutropenia
- Granulocytes frequently left-shifted with occasional immature forms (promyelocytes, myelocytes and metamyelocytes, usually < 10% of the leukocytes)
- Dysplastic features in neutrophils (more frequently related to leukopenia): hyposegmented or abnormally segmented nuclei, abnormal cytoplasmic granulation
- Eosinophilia
- Mild anemia, often normocytic or macrocytic, nucleated red blood cells may be seen
- Thrombocytopenia with atypical, large platelets.
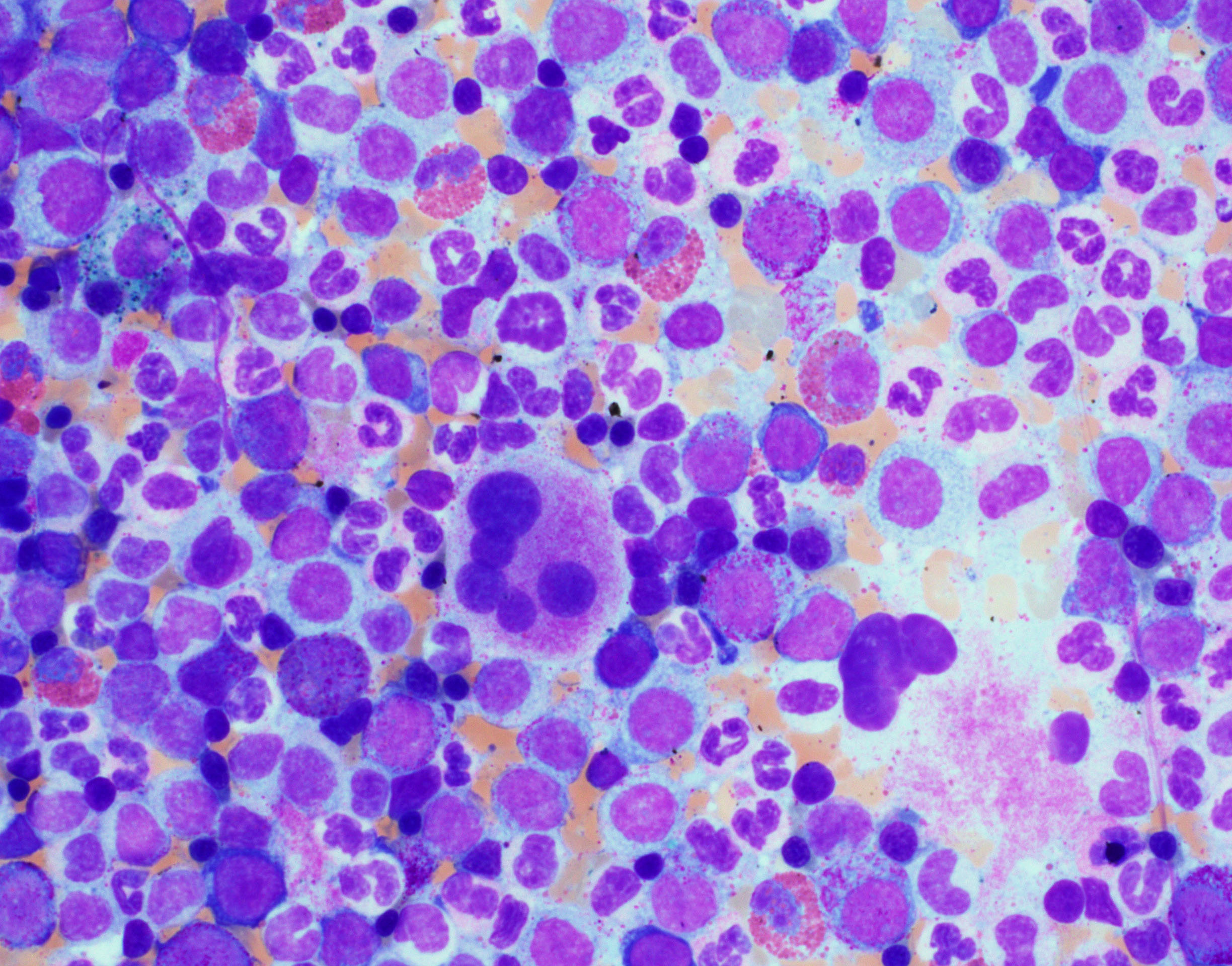
Bone marrow:
- Usually hypercellular (>75% of cases) but rarely can be hypocellular.
- The myeloid to erythroid ratio is generally increased with dysplastic granulocytic hyperplasia.
- Monocytes can be difficult to recognize in the bone marrow, especially when many dysplatic granulocytes are present.
- Cytochemical studies (Alpha-naphthyl butyrate esterase or alpha-naphthyl acetate esterase with fluoride inhibition staining of blood and bone marrow aspirate smears can be useful to facilitate better recognition of the monocytic component.
- Mild dyserythropoiesis (e.g. megaloblastoid changes, abnormal nuclear features and ring sideroblasts) in some cases.
- Dysmegakaryopoiesis are present in many cases.
- Mild to moderate increase in the number of reticulin fibers seen in nearly 30% of cases[8].
- Nodules composed of mature plasmacytoid dendritic cells in the bone marrow biopsy have been reported in 20% of cases[9].
Spleen and lymph node:
- Red pulp infiltrate in spleen.
- Lymph node infiltrate less common
- Involvement by a diffuse infiltration of plasmacytoid dendritic cells can be seen in lymph node and less commonly in spleen
Immunophenotype
The neoplastic monocytic cells frequently show some aberrant immunophenotype. Recognizing monoblasts by immunophenotype can be very challenging.
| Finding | Marker |
|---|---|
| Positive (universal) | CD13, CD33 |
| Positive (variable, can be aberrantly decreased) | CD11c, CD13, CD14, CD15, CD16, CD36, CD64, CD64, CD68, HLA-DR |
| Positive (aberrant, variable) | CD56, CD2. Increased CD34+ blasts suggest disease progression |
| Negative (universal) | CD3, CD5, CD20, CD79a (T-cell and B-cell markers) |
| Negative (subset) | CD14, CD56 |
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.No chromosome rearrangements specifically associated with CMML.
By definition, the presence of BCR-ABL11 or rearrangements involving PDGFRA, PDGFRB or FGFR1 and PCM1-JAK2 defines categories other than CMML.
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
Presence of one or more mutations, especially concurrent mutations in TET2 and SRSF2 is very helpful for a definitive diagnosis.
Survival times range from 1 month to > 100 months; the median survival time in most series is 20-40 months[10][11][12][13].
15- 30% of cases progress to acute myeloid leukemia (AML), classified as AML with myelodysplasia-related changes (MRC).
Important factors predicting progression and survival time[12]:
- Percentage of blasts in blood and bone marrow is the most important factor determining survival
- Karyotype, ASXL1 mutation[14]
- Leukocytosis
Other predictive factors for disease progression:
- Lactate dehydrogenase (LDH) level
- Splenomegaly
- Anemia, thrombocytopenia and lymphocytosis (>2.5 x 109/L)
Somatic ASXL1 mutation has also been incorporated in a clinical prognostic scoring system[14].
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.Clonal cytogenetic abnormalities are found in 20-40% of CMML cases, but there are no specific abnormalities[15].
Chromosomal Abnormality Prevalence Trisomy 8 up to 10% del(20q) up to 10% -Y 4% -7 or del(7q) 1.5% Complex 3%
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.No characteristic chromosome aberrations identified in association with CMML.
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.Over 90% of CMML patients studied exhibited one or more mutations[1]. Concurrent mutations in TET2 and SRSF2 appear to be highly specific for this entity[16][17]. Mutations in certain pathways correlated with clinical classification (dysplastic vs. proliferative)[18].
Gene Mutation Prevalence[1][16] TET2 50-60% SRSF2 40-50% ASXL1 35-40% RUNX1 15-25%, usually secondary[19] NRAS p.G12 most common 11% CBL 10% JAK2 V617F 10% NOTCH 10% Other Mutations
Type Gene/Region/Other Mutation prevalence not well studied in PTPN11, DNMT3A, IDH2, IDH1, SF3B1,U2AF35, ZRSR2, UTX, EZH2, KRAS, CBL, CSF3R, SETBP1
Uncommon Mutations in NPM1, CEBPA, FLT3-ITD, TP53 The clonal evolution and secondary subclonal hierarchy may be correlated with clinical phenotypes or outcomes[18].
Epigenomic Alterations
The genome-wide methylation profiles showed limited applicability in a diagnostic setting[20][21].
The methylation level of individual selected gene promoters have been correlated with overall survival (OS) and progression free survival (PFS)[21]:
- CDH1, ETS1, DAXX, FADD, DAPP1, AATK, CYFIP, TP53, BP2, and AIM2, which were hypermethylated in CMML samples.
- BCL2 hypomethylation is predictive of poorer OS and PFS.
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
Genetic Diagnostic Testing Methods
- Blood cell counts and smear review.
- Bone marrow aspirate and core biopsy with flow cytometric immunophenotyping and immunohistochemical stains.
- Molecular genetic studies are essential to provide clonal evidence for a diagnosis of a myeloid neoplasm, and rule out other well defined entities.
- There are no diagnostic molecular/genetic abnormalities specific for CMML.
Familial Forms
Familial form of CMML has not been reported.
A patient with CMML secondary to familial platelet disorder with germline RUNX1 mutation (classified as myeloid neoplasm with germline RUNX1 mutation) was reported after acquired CBL mutation and 11q-acquired uniparental disomy (11q-aUPD)[22].
Additional Information
Acute myeloid leukemia (AML) with monocytic differentiation frequently have increased circulating mature monocytes (more mature forms than in the bone marrow), the final diagnosis of CML should never be rendered without an evaluation of BM to rule out acute myeloid leukemia. Rarely, CMML can be "marrow-predominant", with monocytes largely confined to the BM without showing absolute monocytosis[23].
Links
Put your text placeholder here (or anywhere appropriate on the page) and use the "Link" icon at the top of the page (Instructions: Highlight text to which you want to add a link in this section or elsewhere, select the "Link" icon at the top of the page, and search the name of the internal page to which you want to link this text, or enter an external internet address by including the "http://www." portion.)
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 1.2 1.3 1.4 Arber DA, et al., (2017). Introduction and overview of the classification of myeloid neoplasms, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Editors. IARC Press: Lyon, France, p82-86.
- ↑ Dinmohamed, Avinash G.; et al. (2015). "The use of medical claims to assess incidence, diagnostic procedures and initial treatment of myelodysplastic syndromes and chronic myelomonocytic leukemia in the Netherlands". Leukemia Research. 39 (2): 177–182. doi:10.1016/j.leukres.2014.11.025.
- ↑ 3.0 3.1 3.2 Czader, M and Orazi, A. Chapter 45. Myelodysplastic/Myeloproliferative Neoplasms and Related Diseases. In Orazi A, Foucar K, Knowles DM. eds. Knowles Neoplastic Hematopathology. Riverwoods, IL: Wolters Kluwer Health; 2013:1140-1156.
- ↑ 4.0 4.1 Germing, Ulrich; et al. (1998). "Problems in the classification of CMML—dysplastic versus proliferative type". Leukemia Research. 22 (10): 871–878. doi:10.1016/S0145-2126(97)00192-6.
- ↑ Onida, Francesco; et al. (2002). "Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients". Blood. 99 (3): 840–849. doi:10.1182/blood.V99.3.840. ISSN 1528-0020.
- ↑ Storniolo AM, Moloney WC, Rosenthal DS, Cox C, Bennett JM. Chronic myelomonocytic leukemia. Leukemia. 1990;4(11):766‐770.
- ↑ Vitte, Franck; et al. (2012). "Specific Skin Lesions in Chronic Myelomonocytic Leukemia: A Spectrum of Myelomonocytic and Dendritic Cell Proliferations. A Study of 42 Cases". The American Journal of Surgical Pathology. 36 (9): 1302–1316. doi:10.1097/PAS.0b013e31825dd4de. ISSN 0147-5185.
- ↑ Ratsimbasoa, Arsène; et al. (2006). "Use of pre-packaged chloroquine for the home management of presumed malaria in Malagasy children". Malaria Journal. 5: 79. doi:10.1186/1475-2875-5-79. ISSN 1475-2875. PMC 1592101. PMID 16972985.
- ↑ Orazi, Attilio; et al. (2006). "Chronic myelomonocytic leukemia: the role of bone marrow biopsy immunohistology". Modern Pathology. 19 (12): 1536–1545. doi:10.1038/modpathol.3800707. ISSN 0893-3952.
- ↑ Longuet, Christine; et al. (2008). "The glucagon receptor is required for the adaptive metabolic response to fasting". Cell metabolism. 8 (5): 359–371. doi:10.1016/j.cmet.2008.09.008. ISSN 1550-4131. PMC 2593715. PMID 19046568.
- ↑ Nijagal, Amar; et al. (2011). "The maternal immune response inhibits the success of in utero hematopoietic cell transplantation". Chimerism. 2 (2): 55–57. doi:10.4161/chim.2.2.16287. ISSN 1938-1956. PMC 3166485. PMID 21912720.
- ↑ 12.0 12.1 U, Germing; et al. (2004). "Risk Assessment in Chronic Myelomonocytic Leukemia (CMML)". PMID 15359628.
- ↑ A, Worsley; et al. (1988). "Prognostic Features of Chronic Myelomonocytic Leukaemia: A Modified Bournemouth Score Gives the Best Prediction of Survival". PMID 3422815.
- ↑ 14.0 14.1 R, Itzykson; et al. (2013). "Prognostic Score Including Gene Mutations in Chronic Myelomonocytic Leukemia". PMID 23690417.
- ↑ Such, E.; et al. (2011). "Cytogenetic risk stratification in chronic myelomonocytic leukemia". Haematologica. 96 (3): 375–383. doi:10.3324/haematol.2010.030957. ISSN 0390-6078. PMC 3046268. PMID 21109693.CS1 maint: PMC format (link)
- ↑ 16.0 16.1 Ti, Mughal; et al. (2015). "An International MDS/MPN Working Group's Perspective and Recommendations on Molecular Pathogenesis, Diagnosis and Clinical Characterization of Myelodysplastic/Myeloproliferative Neoplasms". doi:10.3324/haematol.2014.114660. PMC 4800699. PMID 26341525.CS1 maint: PMC format (link)
- ↑ Gelsi-Boyer, Véronique; et al. (2008). "Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1genes". BMC Cancer. 8 (1). doi:10.1186/1471-2407-8-299. ISSN 1471-2407. PMC 2588460. PMID 18925961.CS1 maint: PMC format (link)
- ↑ 18.0 18.1 Patel, B J; et al. (2017). "Genomic determinants of chronic myelomonocytic leukemia". Leukemia. 31 (12): 2815–2823. doi:10.1038/leu.2017.164. ISSN 0887-6924.
- ↑ Palomo, Laura; et al. (2020). "Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms". Blood. doi:10.1182/blood.2019004229. ISSN 0006-4971.
- ↑ Deininger, Michael W.; et al. (2018). "Molecular Alterations in Chronic Myelomonocytic Leukemia Monocytes: Transcriptional and Methylation Profiling". Blood. 132 (Supplement 1): 3889–3889. doi:10.1182/blood-2018-99-115077. ISSN 0006-4971.
- ↑ 21.0 21.1 Palomo, Laura; et al. (2018). "DNA methylation profile in chronic myelomonocytic leukemia associates with distinct clinical, biological and genetic features". Epigenetics. 13 (1): 8–18. doi:10.1080/15592294.2017.1405199. ISSN 1559-2294. PMC 5837079. PMID 29160764.CS1 maint: PMC format (link)
- ↑ Shiba, Norio; et al. (2012). "CBL mutation in chronic myelomonocytic leukemia secondary to familial platelet disorder with propensity to develop acute myeloid leukemia (FPD/AML)". Blood. 119 (11): 2612–2614. doi:10.1182/blood-2011-02-333435. ISSN 0006-4971.
- ↑ A, Orazi; et al. (2008). "The Myelodysplastic/Myeloproliferative Neoplasms: Myeloproliferative Diseases With Dysplastic Features". PMID 18480833.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Chronic myelomonocytic leukaemia”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 09/6/2024, https://ccga.io/index.php/HAEM5:Chronic_myelomonocytic_leukaemia.