Difference between revisions of "HAEM5:Myeloid/lymphoid neoplasm with PDGFRA rearrangement"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) (Created page with "{{DISPLAYTITLE:Myeloid/lymphoid neoplasm with PDGFRA rearrangement}} Haematolymphoid Tumours (5th ed.) {{Under Construction}} <blockquote class='...") |
Bailey.Glen (talk | contribs) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Myeloid/lymphoid neoplasm with PDGFRA rearrangement}} | {{DISPLAYTITLE:Myeloid/lymphoid neoplasm with PDGFRA rearrangement}} | ||
| − | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] |
{{Under Construction}} | {{Under Construction}} | ||
| − | <blockquote class='blockedit'>{{Box-round|title= | + | <blockquote class='blockedit'>{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Myeloid/Lymphoid Neoplasms with PDGFRA Rearrangement]]. |
}}</blockquote> | }}</blockquote> | ||
| + | |||
| + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> | ||
| + | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| Line 12: | Line 15: | ||
__TOC__ | __TOC__ | ||
| − | == | + | ==WHO Classification of Disease== |
| − | + | {| class="wikitable" | |
| − | + | !Structure | |
| − | + | !Disease | |
| − | + | |- | |
| − | Myeloid/lymphoid neoplasms with eosinophilia and gene rearrangement | + | |Book |
| + | |Haematolymphoid Tumours (5th ed.) | ||
| + | |- | ||
| + | |Category | ||
| + | |Myeloid proliferations and neoplasms | ||
| + | |- | ||
| + | |Family | ||
| + | |Myeloid/lymphoid neoplasms | ||
| + | |- | ||
| + | |Type | ||
| + | |Myeloid/lymphoid neoplasms with eosinophilia and defining gene rearrangement | ||
| + | |- | ||
| + | |Subtype(s) | ||
| + | |Myeloid/lymphoid neoplasm with PDGFRA rearrangement | ||
| + | |} | ||
==Definition / Description of Disease== | ==Definition / Description of Disease== | ||
| Line 44: | Line 61: | ||
==Clinical Features== | ==Clinical Features== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> |
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |EXAMPLE Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | EXAMPLE B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | EXAMPLE Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | EXAMPLE Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |EXAMPLE Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | EXAMPLE Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 117: | Line 134: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 177: | Line 194: | ||
</blockquote> | </blockquote> | ||
| − | ==Individual Region Genomic Gain/Loss/LOH== | + | ==Individual Region Genomic Gain / Loss / LOH== |
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable.'') </span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 189: | Line 206: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |EXAMPLE Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 202: | Line 219: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |EXAMPLE Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 219: | Line 236: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 225: | Line 242: | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| − | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis'')</span> | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 235: | Line 252: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 241: | Line 258: | ||
|No | |No | ||
|No | |No | ||
| − | |EXAMPLE: | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 251: | Line 268: | ||
</blockquote> | </blockquote> | ||
| − | ==Gene Mutations (SNV/INDEL)== | + | ==Gene Mutations (SNV / INDEL)== |
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 263: | Line 280: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |EXAMPLE: TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | EXAMPLE: | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | EXAMPLE: BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |EXAMPLE: TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |EXAMPLE: 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | EXAMPLE: 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |EXAMPLE: IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |EXAMPLE: EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 295: | Line 312: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table.'')</span> | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |EXAMPLE: BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |EXAMPLE: MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |EXAMPLE: Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |EXAMPLE: CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |EXAMPLE: Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |EXAMPLE: Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |EXAMPLE: KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |EXAMPLE: Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |EXAMPLE: Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
Latest revision as of 17:21, 6 September 2024
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Myeloid/Lymphoid Neoplasms with PDGFRA Rearrangement.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Jay Alden, DO
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Myeloid/lymphoid neoplasms |
| Type | Myeloid/lymphoid neoplasms with eosinophilia and defining gene rearrangement |
| Subtype(s) | Myeloid/lymphoid neoplasm with PDGFRA rearrangement |
Definition / Description of Disease
Myeloproliferative neoplasms associated with PDGFRA rearrangements are primary/neoplastic hypereosinophilic syndromes associated with recurrent rearrangements of the PDGFRA gene [1]. It is most commonly associated with FIP1L1-PDGFRA (F/P) fusion resulting from a cryptic deletion at 4q12, [2] and often presents as chronic eosinophilic leukemia (CEL), or less commonly, acute myeloid leukemia or T-lymphoblastic leukemia/lymphoma. [3]
Synonyms / Terminology
Chronic eosinophilic leukemia with FIP1L1-PDGFRA
FIP1L1-PDGFRA –associated chronic eosinophilic leukemia
Myeloid and lymphoid neoplasms associated with PDGFRA rearrangement
PDGFRA-associated Hypereosinophilic syndrome
Myeloid and lymphoid neoplasms with PDGFRA rearrangement
Myeloproliferative variant of the hypereosinophilic syndrome [4]
Epidemiology / Prevalence
The incidence and prevalence of myeloid/lymphoid neoplasms with PDGFRA rearrangement is not well characterized as demographic data is scarce [5]. The incidence of HES of any cause is estimated at 0.036 per 100,000, [6] though cases in which a causative genetic abnormality constitute a minority of these cases [5]. The F/P rearrangement is the most common abnormality identified, and is estimated to comprise approximately 10% of patients with significant hypereosinophilia [7] [8]. The entity is recently described, and disorders once called idiopathic hypereosinophilic syndrome are now being classified with genetic testing as specific primary neoplasms or reactive conditions. The F/P rearrangment is significantly more common in males with a male:female ratio of about 17:1. The age range varies from ages 7-77 with most patients being between 25 and 55 years. [9]
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.Clinical presentation ranges from asymptomatic to fulminant, life threatening multi-system organ failure. Presenting signs and symptoms are typically related to eosinophilic infiltration, and consistent with hypereosinophilic syndromes of any cause. The largest clinical analysis of patients with hypereosinophilic syndromes (HES) demonstrated the following manifestations at presentation:[10]
- Dermatologic (eg, rash) – 57 percent
- Pulmonary (cough and breathlessness) – 25 percent
- Gastrointestinal – 14 percent
- Cardiac – <5 percent
- Asymptomatic -- 6 percent
Neoplastic PDGFRA-associated hypereosinophilic syndromes are more likely to present with eosinophilic cardiopulmonary disease than HES of any cause. A survey of 44 cases demonstrated skin, spleen, lung, and heart involvement in 57, 52, 45, and 34 percent of cases respectively with a similar rate of asymptomatic cases. [11]
Sites of Involvement
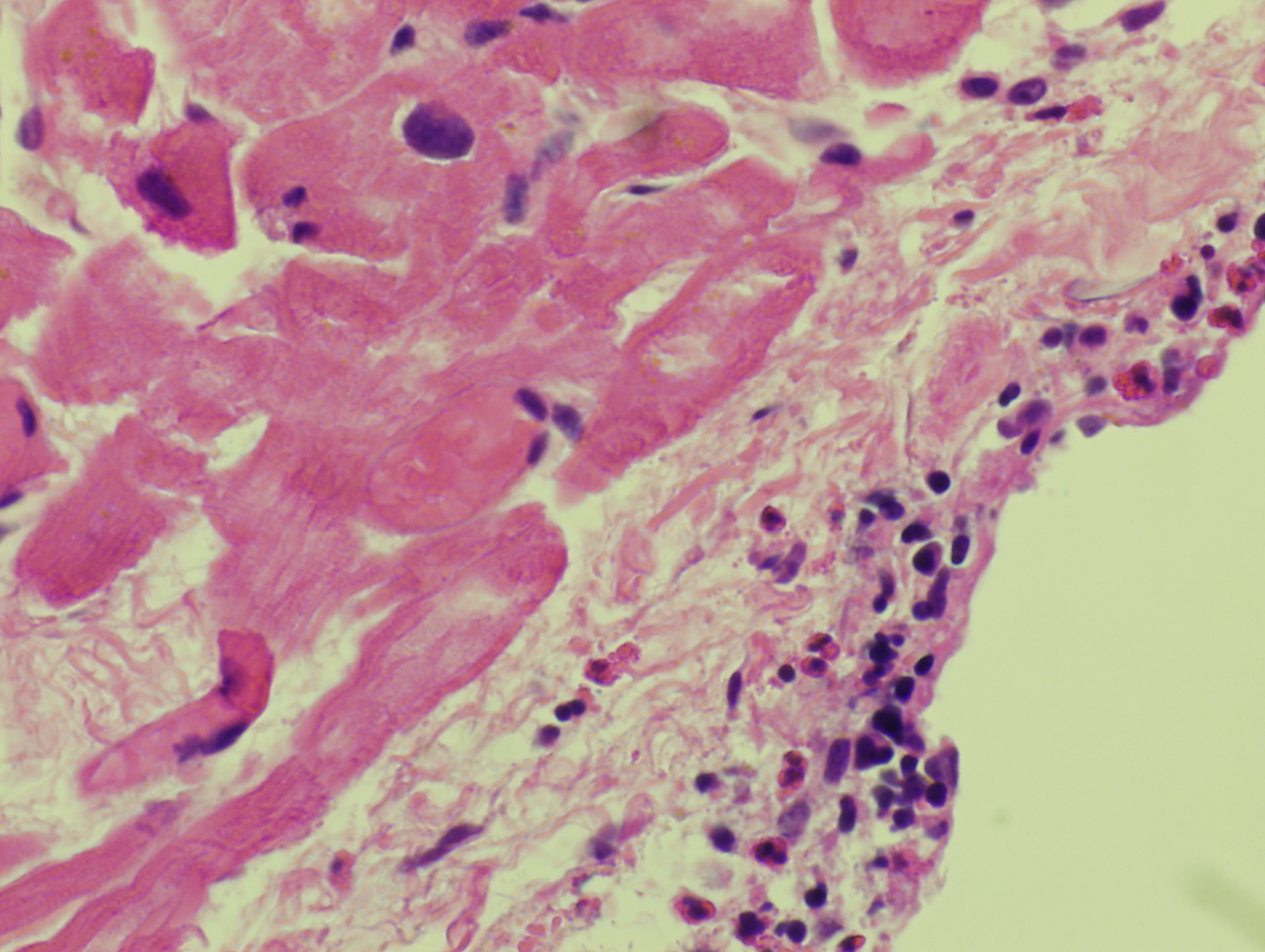
Leukemia associated with F/P is a systemic disease occupying the bone marrow and peripheral blood. Neoplastic cells may infiltrate various tissues such as the heart, lungs, nervous systems, skin and GI tract resulting in degranulation and cytokine mediated injury. [1]
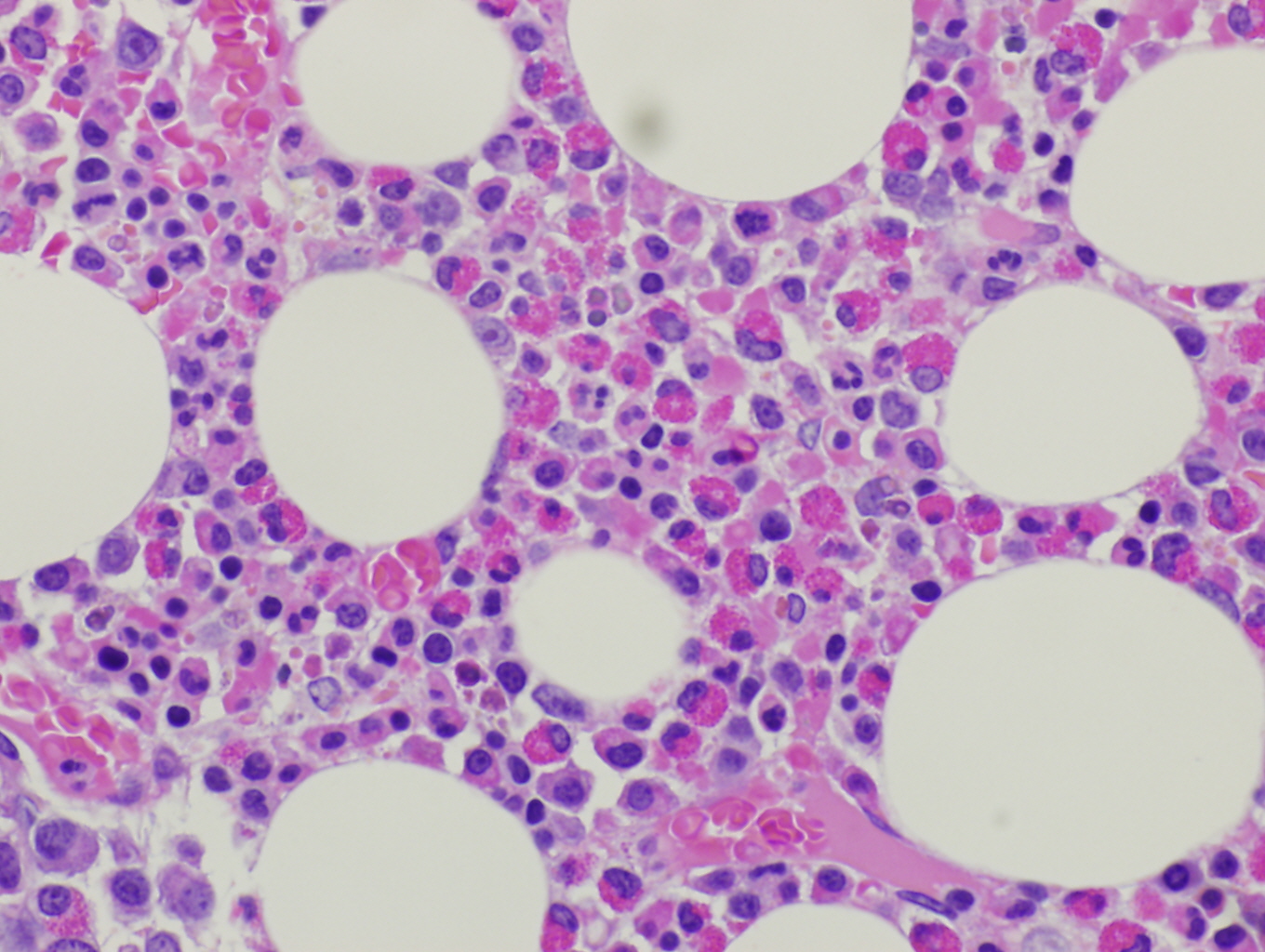
Morphologic Features
Histopathologic features are dependent on the organs involved. Eosinophilic infiltration is noted on diagnostic tissue biopsy. Eosinophilia and increased eosinophilic precursors can be identified on trephine bone marrow biopsy and peripheral blood smears. There is no histologic correlate to clonality, and genetic studies are required for diagnosis.
Immunophenotype
These neoplastic eosinophils may express markers of activation such as CD23, CD25, and CD69[4]. The basophils can sometimes be distinguished from those in systemic mastocytosis as CD2 is typically negative in the mast cells of PDGFRA rearrangement, but positive in systemic mastocytosis. [12][3]
| Finding | Marker |
|---|---|
| Positive (subset) | CD25 [4] |
| Positive (subset) | CD23 [4] |
| Positive (subset) | CD69 [4] |
| Negative (Mast cells) | CD2 [12][3] |
| Posivite (Mast cells, subset | CD25 [12][3] |
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.
Chromosomal Rearrangement Genes in Fusion (5’ or 3’ Segments) Prevalence Cryptic del(4)(q12) FIP1L1-PDGFRA Majority T(1;4) (q44;q12) FIP1L1-PDGFRA Rare [13] T(4;10)(q12;p11.1-p11.2) FIP1L1-PDGFRA Rare [14] T(4;22)(q12;q11.2) BCR-PDGFRA At least 9 cases [15][16][17][18] T(2;4)(p24;q12) STRN-PDGFRA At least 1 case [19] T(4;12)(q12;p13.2) ETV6-PDGFRA At least 1 case [19] ins(9;4)(q33;q12q25) CDK5RAP2-PDGFRA At least 1 case [20] T(4;10)(q12;q23.3) TNKS2-PDGFRA At least 1 case [21]
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
The responsiveness of F/P associated myeloid/lymphoid neoplasms to imatinib mesylate is well documented [5]. Adverse outcomes are typically related to late presentation, where irreversible organ damage precedes diagnosis, or when the disease is diagnosed in an accelerated phase when complications are more likely. Induction dosing of imatinib ranges from 100-400 mg daily, with much lower maintenence dosing recommended to prevent relapse [22] [23]. Complete hematologic and molecular remission is observed in nearly all patients taking imatinib, usually within 3 months. [24] [25] [26] Imatinib maintains efficacy in accelerated or blast phase disease, and resistance is rare [11] [27].
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.Cytogenetic studies are usually normal though trisomy of chromosome 8 has been described, and may represent disease evolution [1].
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.An activating point mutation in PDGFRA has also been described [28].
Epigenomic Alterations
Put your text here
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.the F/P tyrosine kinase is thought to become constitutively active in the setting of PDGRA juxtamembrane interruption as breakpoints in the PDGRA gene are tightly clustered, resulting in the removal of a portion of the juxtamembrane domain and activation of the kinase domain upon rearrangement. The role of the FIP1L1 in the neoplastic process is thought to be less significant. [29] The eosinophilic proliferation observed in these patients is thought to result from multiple signalling pathways including phosphoinositol 3-kinase, ERK 1/2 and STAT5, though the precise mechanism remains elusive. [2][30]
Genetic Diagnostic Testing Methods
Myeloid/lymphoid neoplasms with PDGFRA are diagnosed with a combination of morphologic, immunophenotypic and genomic modalities; typically with a bone marrow biopsy and peripheral blood smear review, preferably drawn prior to administration of high dose corticosteroids. Additional studies performed at the time of workup include CBC to quantify any abnormalities in other cell lines, tests of hepatic and renal function, troponin for evidence of myocarditis, vitamin B12, serum tryptase, antineutrophil cytoplasmic antibodies, and a high resolution chest CT for evidence of pulmonary involvement. [31] The diagnosis is made when the F/P fusion gene or a variant fusion gene with rearrangement of PDGFRA or an activating mutation of PDGFRA is identified in the setting of a myeloid or lymphoid neoplasm, usually with prominent eosinophilia [1].
The F/P fusion gene can be detected by reverse transcriptase PCR (RT-PCR) [13], or the deletion can be detected using a probe for the CHIC2 gene, or with a break apart probe encompassing FIP1L1 and PDGFRA. [1]
Familial Forms
Put your text here (Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.)
Additional Information
Put your text here
Links
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 1.2 1.3 1.4 Bain BJ, et al., (2017). Myeloid/lymphoid neoplasms with PDGFRA rearrangement in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France, p73-75.
- ↑ 2.0 2.1 Cools, Jan; et al. (2003). "A Tyrosine Kinase Created by Fusion of the PDGFRA and FIP1L1 Genes as a Therapeutic Target of Imatinib in Idiopathic Hypereosinophilic Syndrome". New England Journal of Medicine. 348 (13): 1201–1214. doi:10.1056/NEJMoa025217. ISSN 0028-4793.
- ↑ 3.0 3.1 3.2 3.3 Metzgeroth, G.; et al. (2007). "Recurrent finding of the FIP1L1-PDGFRA fusion gene in eosinophilia-associated acute myeloid leukemia and lymphoblastic T-cell lymphoma". Leukemia. 21 (6): 1183–1188. doi:10.1038/sj.leu.2404662. ISSN 0887-6924. PMID 17377585.
- ↑ 4.0 4.1 4.2 4.3 4.4 Klion, Amy D.; et al. (2004). "Molecular remission and reversal of myelofibrosis in response to imatinib mesylate treatment in patients with the myeloproliferative variant of hypereosinophilic syndrome". Blood. 103 (2): 473–478. doi:10.1182/blood-2003-08-2798. ISSN 0006-4971. PMID 14504092.
- ↑ 5.0 5.1 5.2 Shomali, William; et al. (2019). "World Health Organization-defined eosinophilic disorders: 2019 update on diagnosis, risk stratification, and management". American Journal of Hematology. 94 (10): 1149–1167. doi:10.1002/ajh.25617. ISSN 1096-8652. PMID 31423623.
- ↑ Crane, Martin M.; et al. (2010). "Incidence of myeloproliferative hypereosinophilic syndrome in the United States and an estimate of all hypereosinophilic syndrome incidence". The Journal of Allergy and Clinical Immunology. 126 (1): 179–181. doi:10.1016/j.jaci.2010.03.035. ISSN 1097-6825. PMC 5781228. PMID 20639012.
- ↑ Pardanani, Animesh; et al. (2004). "FIP1L1-PDGFRA fusion: prevalence and clinicopathologic correlates in 89 consecutive patients with moderate to severe eosinophilia". Blood. 104 (10): 3038–3045. doi:10.1182/blood-2004-03-0787. ISSN 0006-4971. PMID 15284118.
- ↑ Pardanani, A.; et al. (2006). "FIP1L1-PDGFRA in eosinophilic disorders: prevalence in routine clinical practice, long-term experience with imatinib therapy, and a critical review of the literature". Leukemia Research. 30 (8): 965–970. doi:10.1016/j.leukres.2005.11.011. ISSN 0145-2126. PMID 16406016.
- ↑ Bain, Barbara J.; et al. (2007). "Chronic eosinophilic leukemias and the myeloproliferative variant of the hypereosinophilic syndrome". Immunology and Allergy Clinics of North America. 27 (3): 377–388. doi:10.1016/j.iac.2007.06.001. ISSN 0889-8561. PMID 17868855.
- ↑ Ogbogu, Princess U.; et al. (2009). "Hypereosinophilic syndrome: a multicenter, retrospective analysis of clinical characteristics and response to therapy". The Journal of Allergy and Clinical Immunology. 124 (6): 1319–1325.e3. doi:10.1016/j.jaci.2009.09.022. ISSN 1097-6825. PMC 2829669. PMID 19910029.
- ↑ 11.0 11.1 Legrand, Fanny; et al. (2013). "The Spectrum of FIP1L1-PDGFRA-Associated Chronic Eosinophilic Leukemia: New Insights Based on a Survey of 44 Cases". Medicine. 92 (5): e1–e9. doi:10.1097/MD.0b013e3182a71eba. ISSN 1536-5964. PMC 4553979. PMID 23982058.
- ↑ 12.0 12.1 12.2 Klion, Amy D.; et al. (2003). "Elevated serum tryptase levels identify a subset of patients with a myeloproliferative variant of idiopathic hypereosinophilic syndrome associated with tissue fibrosis, poor prognosis, and imatinib responsiveness". Blood. 101 (12): 4660–4666. doi:10.1182/blood-2003-01-0006. ISSN 0006-4971. PMID 12676775.
- ↑ 13.0 13.1 Cools, Jan; et al. (2003). "A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome". The New England Journal of Medicine. 348 (13): 1201–1214. doi:10.1056/NEJMoa025217. ISSN 1533-4406. PMID 12660384.
- ↑ Tashiro, Haruko; et al. (2006). "Molecular analysis of chronic eosinophilic leukemia with t(4;10) showing good response to imatinib mesylate". International Journal of Hematology. 83 (5): 433–438. doi:10.1532/IJH97.05180. ISSN 0925-5710. PMID 16787876.
- ↑ Baxter, E. Joanna; et al. (2002). "The t(4;22)(q12;q11) in atypical chronic myeloid leukaemia fuses BCR to PDGFRA". Human Molecular Genetics. 11 (12): 1391–1397. doi:10.1093/hmg/11.12.1391. ISSN 0964-6906. PMID 12023981.
- ↑ Van Etten, Richard A.; et al. (2006). "Distinct Leukemogenic Activity and Imatinib Responsiveness of a BCR-PFGFRα Fusion Tyrosine Kinase". Blood. 108 (11): 3634–3634. doi:10.1182/blood.V108.11.3634.3634. ISSN 0006-4971.
- ↑ Safley, Anne Michele; et al. (2004). "Molecular and cytogenetic characterization of a novel translocation t(4;22) involving the breakpoint cluster region and platelet-derived growth factor receptor-alpha genes in a patient with atypical chronic myeloid leukemia". Genes, Chromosomes & Cancer. 40 (1): 44–50. doi:10.1002/gcc.20014. ISSN 1045-2257. PMID 15034867.
- ↑ Trempat, Pascal; et al. (2003). "Chronic myeloproliferative disorders with rearrangement of the platelet-derived growth factor alpha receptor: a new clinical target for STI571/Glivec". Oncogene. 22 (36): 5702–5706. doi:10.1038/sj.onc.1206543. ISSN 0950-9232. PMID 12944919.
- ↑ 19.0 19.1 Curtis, Claire E.; et al. (2007). "Two novel imatinib-responsive PDGFRA fusion genes in chronic eosinophilic leukaemia". British Journal of Haematology. 138 (1): 77–81. doi:10.1111/j.1365-2141.2007.06628.x. ISSN 0007-1048. PMID 17555450.
- ↑ Walz, Christoph; et al. (2006). "Transient response to imatinib in a chronic eosinophilic leukemia associated with ins(9;4)(q33;q12q25) and a CDK5RAP2-PDGFRA fusion gene". Genes, Chromosomes & Cancer. 45 (10): 950–956. doi:10.1002/gcc.20359. ISSN 1045-2257. PMID 16845659.
- ↑ Chalmers, Z. R.; et al. (2015). "Comprehensive genomic profiling identifies a novel TNKS2-PDGFRA fusion that defines a myeloid neoplasm with eosinophilia that responded dramatically to imatinib therapy". Blood Cancer Journal. 5: e278. doi:10.1038/bcj.2014.95. ISSN 2044-5385. PMC 4349257. PMID 25658984.
- ↑ Baccarani, Michele; et al. (2007). "The efficacy of imatinib mesylate in patients with FIP1L1-PDGFRalpha-positive hypereosinophilic syndrome. Results of a multicenter prospective study". Haematologica. 92 (9): 1173–1179. doi:10.3324/haematol.11420. ISSN 1592-8721. PMID 17666373.
- ↑ Jovanovic, Jelena V.; et al. (2007). "Low-dose imatinib mesylate leads to rapid induction of major molecular responses and achievement of complete molecular remission in FIP1L1-PDGFRA-positive chronic eosinophilic leukemia". Blood. 109 (11): 4635–4640. doi:10.1182/blood-2006-10-050054. ISSN 0006-4971. PMID 17299092.
- ↑ Baccarani, Michele; et al. (2007). "The efficacy of imatinib mesylate in patients with FIP1L1-PDGFRalpha-positive hypereosinophilic syndrome. Results of a multicenter prospective study". Haematologica. 92 (9): 1173–1179. doi:10.3324/haematol.11420. ISSN 1592-8721. PMID 17666373.
- ↑ Quéméneur, Thomas; et al. (2013). "Systemic vasculitis during the course of systemic sclerosis: report of 12 cases and review of the literature". Medicine. 92 (1): 1–9. doi:10.1097/MD.0b013e31827781fd. ISSN 1536-5964. PMC 5370746. PMID 23263715.
- ↑ Helbig, Grzegorz; et al. (2008). "A single weekly dose of imatinib is sufficient to induce and maintain remission of chronic eosinophilic leukaemia in FIP1L1-PDGFRA-expressing patients". British Journal of Haematology. 141 (2): 200–204. doi:10.1111/j.1365-2141.2008.07033.x. ISSN 1365-2141. PMID 18307562.
- ↑ Lierman, E.; et al. (2009). "FIP1L1-PDGFRalpha D842V, a novel panresistant mutant, emerging after treatment of FIP1L1-PDGFRalpha T674I eosinophilic leukemia with single agent sorafenib". Leukemia. 23 (5): 845–851. doi:10.1038/leu.2009.2. ISSN 1476-5551. PMID 19212337.
- ↑ Elling, Christian; et al. (2011). "Novel imatinib-sensitive PDGFRA-activating point mutations in hypereosinophilic syndrome induce growth factor independence and leukemia-like disease". Blood. 117 (10): 2935–2943. doi:10.1182/blood-2010-05-286757. ISSN 1528-0020. PMID 21224473.
- ↑ J. Cools; et al. (2008). "Five years since the discovery of FIP1L1–PDGFRA : what we have learned about the fusion and other molecularly defined eosinophilias". Leukemia. 22 (11): 1999–2010. doi:10.1038/leu.2008.287. ISSN 1476-5551.
- ↑ Buitenhuis, Miranda; et al. (2007). "Molecular mechanisms underlying FIP1L1-PDGFRA-mediated myeloproliferation". Cancer Research. 67 (8): 3759–3766. doi:10.1158/0008-5472.CAN-06-4183. ISSN 0008-5472. PMID 17440089.
- ↑ Rosemarin, A, and Feldweg, A. Hypereosinophilic syndromes: Treatement. In: UpToDate, Post, TW (Ed), UpToDate, Waltham, MA. April 2020
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. *Citation of this Page: “Myeloid/lymphoid neoplasm with PDGFRA rearrangement”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 09/6/2024, https://ccga.io/index.php/HAEM5:Myeloid/lymphoid_neoplasm_with_PDGFRA_rearrangement.