Difference between revisions of "BRST5:Secretory carcinoma"
| [unchecked revision] | [checked revision] |
Kgeiersbach (talk | contribs) (added gene fusion diagram) |
|||
| (15 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | {{DISPLAYTITLE:Secretory carcinoma}} |
| + | |||
| + | [[BRST5:Table_of_Contents|Breast Tumours (WHO Classification, 5th ed.)]] | ||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
| + | Hui Chen, MD, PhD, The University of Texas MD Anderson Cancer Center, Houston, TX, USA | ||
| − | + | Katherine Geiersbach, MD, Mayo Clinic - Rochester, MN, USA | |
| − | + | ==WHO Classification of Disease== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
| − | + | !Structure | |
| − | + | !Disease | |
|- | |- | ||
| − | | | + | |Book |
| − | | | + | |Breast Tumours (5th ed.) |
| + | |- | ||
| + | |Category | ||
| + | |Epithelial tumours of the breast | ||
| + | |- | ||
| + | |Family | ||
| + | |Rare and salivary gland-type tumours: Introduction | ||
| + | |- | ||
| + | |Type | ||
| + | |Secretory carcinoma | ||
| + | |- | ||
| + | |Subtype(s) | ||
| + | |N/A | ||
|} | |} | ||
| − | == | + | ==WHO Essential and Desirable Genetic Diagnostic Criteria== |
| − | + | {| class="wikitable" | |
| − | + | |+ | |
| − | + | |WHO Essential Criteria (Genetics)* | |
| − | + | | | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | {| class="wikitable | ||
|- | |- | ||
| − | + | |WHO Desirable Criteria (Genetics)* | |
| + | |''ETV6''::''NTRK3'' fusion | ||
|- | |- | ||
| − | | | + | |Other Classification |
| + | | | ||
| + | |} | ||
| + | <nowiki>*</nowiki>Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the [https://tumourclassification.iarc.who.int/home <u>WHO Classification of Tumours</u>]. | ||
| + | ==Related Terminology== | ||
| + | {| class="wikitable" | ||
| + | |+ | ||
| + | |Acceptable | ||
| + | | | ||
|- | |- | ||
| − | | | + | |Not Recommended |
| − | + | | | |
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | == | + | ==Gene Rearrangements== |
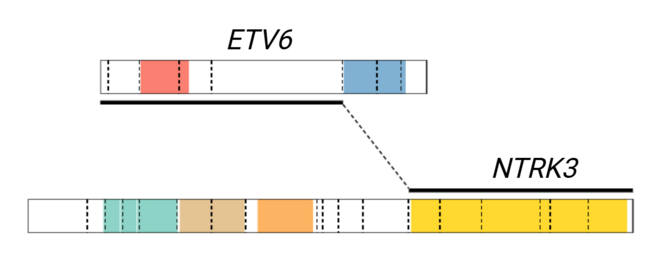
| − | + | [[File:ETV6-NTRK3 fusion diagram.tif|left|frameless|655x655px|Gene fusion diagram showing the canonical breakpoints in exon 5 of ''ETV6'' (NM_001987) and exon 15 of ''NTRK3'' (NM_001012338). Alternate fusion breakpoints include exon 4 of ''ETV6'' and exon 14 of ''NTRK3''.]] | |
| − | + | <br /> | |
| − | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| − | ! | + | !Driver Gene!!Fusion(s) and Common Partner Genes!!Molecular Pathogenesis!!Typical Chromosomal Alteration(s) |
| − | ! | + | !Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
| − | !Prognostic Significance | + | !Diagnostic, Prognostic, and Therapeutic Significance - D, P, T |
| − | ! | + | !Established Clinical Significance Per Guidelines - Yes or No (Source) |
| − | !Notes | + | !Clinical Relevance Details/Other Notes |
| + | |- | ||
| + | |''NTRK3'' | ||
| + | |''ETV6''::''NTRK3''<ref>{{Cite journal|last=Tognon|first=Cristina|last2=Knezevich|first2=Stevan R.|last3=Huntsman|first3=David|last4=Roskelley|first4=Calvin D.|last5=Melnyk|first5=Natalya|last6=Mathers|first6=Joan A.|last7=Becker|first7=Laurence|last8=Carneiro|first8=Fatima|last9=MacPherson|first9=Nicol|date=2002-11|title=Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma|url=https://pubmed.ncbi.nlm.nih.gov/12450792|journal=Cancer Cell|volume=2|issue=5|pages=367–376|doi=10.1016/s1535-6108(02)00180-0|issn=1535-6108|pmid=12450792}}</ref> | ||
| + | |Fusion results in constitutive activation of NTRK3 tyrosine kinase | ||
| + | |t(12;15)(p13;q25) | ||
| + | |Common | ||
| + | |D, P, T | ||
| + | | | ||
| + | |The ''ETV6''::''NTRK3'' fusion is diagnostic of secretory carcinoma in the appropriate morphologic and clinical context.<ref>{{Cite journal|last=Arce|first=C.|last2=Cortes-Padilla|first2=D.|last3=Huntsman|first3=D. G.|last4=Miller|first4=M. A.|last5=Dueñnas-Gonzalez|first5=A.|last6=Alvarado|first6=A.|last7=Pérez|first7=V.|last8=Gallardo-Rincón|first8=D.|last9=Lara-Medina|first9=F.|date=2005-06-17|title=Secretory carcinoma of the breast containing the ETV6-NTRK3 fusion gene in a male: case report and review of the literature|url=https://pubmed.ncbi.nlm.nih.gov/15963235|journal=World Journal of Surgical Oncology|volume=3|pages=35|doi=10.1186/1477-7819-3-35|issn=1477-7819|pmc=1184104|pmid=15963235}}</ref><ref>{{Cite journal|last=Jacob|first=John Doromal|last2=Hodge|first2=Caitlin|last3=Franko|first3=Jan|last4=Pezzi|first4=Christopher M.|last5=Goldman|first5=Charles D.|last6=Klimberg|first6=Vicki Suzanne|date=2016-06|title=Rare breast cancer: 246 invasive secretory carcinomas from the National Cancer Data Base|url=https://pubmed.ncbi.nlm.nih.gov/27040042|journal=Journal of Surgical Oncology|volume=113|issue=7|pages=721–725|doi=10.1002/jso.24241|issn=1096-9098|pmid=27040042}}</ref><ref>{{Cite journal|last=Li|first=Dali|last2=Xiao|first2=Xiuying|last3=Yang|first3=Wentao|last4=Shui|first4=Ruohong|last5=Tu|first5=Xiaoyu|last6=Lu|first6=Hongfen|last7=Shi|first7=Daren|date=2012-04|title=Secretory breast carcinoma: a clinicopathological and immunophenotypic study of 15 cases with a review of the literature|url=https://pubmed.ncbi.nlm.nih.gov/22157932|journal=Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc|volume=25|issue=4|pages=567–575|doi=10.1038/modpathol.2011.190|issn=1530-0285|pmid=22157932}}</ref> This fusion is responsive to TRK inhibitor therapies such as larotrectinib abd entrectinib. | ||
|- | |- | ||
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | + | | | |
| + | | | ||
| + | | | ||
| + | | | ||
| + | |} | ||
| + | |||
| − | |||
| − | |||
| − | |||
==Individual Region Genomic Gain/Loss/LOH== | ==Individual Region Genomic Gain/Loss/LOH== | ||
| − | + | <br /> | |
| − | |||
| − | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| − | !Chr #!!Gain | + | !Chr #!!'''Gain, Loss, Amp, LOH'''!!'''Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size]'''!!'''Relevant Gene(s)''' |
| − | !Diagnostic | + | !'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' |
| − | + | !'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | |
| − | ! | + | !'''Clinical Relevance Details/Other Notes''' |
| − | !Notes | ||
|- | |- | ||
| − | | | + | | |
| − | + | | | |
| − | + | | | |
| − | + | | | |
| − | + | | | |
| − | + | | | |
| − | + | | | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | | | ||
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | |||
| − | | | ||
| − | | | ||
| − | | | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | |||
| − | |||
| + | ==Characteristic Chromosomal or Other Global Mutational Patterns== | ||
| + | <br /> | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Chromosomal Pattern | !Chromosomal Pattern | ||
| − | ! | + | !Molecular Pathogenesis |
| − | !Prognostic Significance | + | !'''Prevalence -''' |
| − | ! | + | '''Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' |
| − | !Notes | + | !'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T''' |
| + | !'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | ||
| + | !'''Clinical Relevance Details/Other Notes''' | ||
|- | |- | ||
| − | | | + | | |
| − | + | | | |
| − | + | | | |
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | | | ||
| − | |||
| − | |||
|} | |} | ||
| − | |||
| − | |||
| + | ==Gene Mutations (SNV/INDEL) == | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| − | !Gene | + | !Gene!!'''Genetic Alteration'''!!'''Tumor Suppressor Gene, Oncogene, Other'''!!'''Prevalence -''' |
| − | !''' | + | '''Common >20%, Recurrent 5-20% or Rare <5% (Disease)''' |
| − | ! | + | !'''Diagnostic, Prognostic, and Therapeutic Significance - D, P, T ''' |
| − | + | !'''Established Clinical Significance Per Guidelines - Yes or No (Source)''' | |
| − | + | !'''Clinical Relevance Details/Other Notes''' | |
|- | |- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | | | ||
| | | | ||
| | | | ||
| − | | | + | | |
| − | + | | | |
| − | |} | + | | |
| − | Note: A more extensive list of mutations can be found in | + | | |
| + | |}Note: A more extensive list of mutations can be found in [https://www.cbioportal.org/ <u>cBioportal</u>], [https://cancer.sanger.ac.uk/cosmic <u>COSMIC</u>], and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | ||
| + | |||
==Epigenomic Alterations== | ==Epigenomic Alterations== | ||
| − | + | <br /> | |
| − | |||
| − | |||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
| − | + | <br /> | |
| − | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | | | + | |''NTRK3''; Activating fusion with 5' partner ''ETV6'' |
| − | | | + | |MAPK/PI3K/AKT signaling |
| − | | | + | |Increased cell growth and proliferation |
|- | |- | ||
| − | | | + | | |
| − | | | + | | |
| − | | | + | | |
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | |||
| − | |||
| + | ==Genetic Diagnostic Testing Methods== | ||
| + | FISH, RT-PCR, next generation sequencing | ||
==Familial Forms== | ==Familial Forms== | ||
| − | + | None | |
| − | |||
| − | |||
==Additional Information== | ==Additional Information== | ||
| − | + | <br /> | |
| − | |||
| − | |||
==Links== | ==Links== | ||
| + | https://www.pathologyoutlines.com/topic/breastmalignantjuvenile.html | ||
| − | + | <br /> | |
| + | ==Notes== | ||
| − | ==References== | + | Prior Author(s): |
| + | ==References == | ||
<references /> | <references /> | ||
| − | + | <nowiki>*</nowiki>''Citation of this Page'': “Secretory carcinoma”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/BRST5:Secretory carcinoma</nowiki>. | |
| − | + | [[Category:BRST5]] | |
| − | + | [[Category:DISEASE]] | |
| − | + | [[Category:Diseases S]] | |
| − | |||
| − | |||
| − | <nowiki>*</nowiki> | ||
Latest revision as of 11:32, 16 April 2025
Breast Tumours (WHO Classification, 5th ed.)
Primary Author(s)*
Hui Chen, MD, PhD, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Katherine Geiersbach, MD, Mayo Clinic - Rochester, MN, USA
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Breast Tumours (5th ed.) |
| Category | Epithelial tumours of the breast |
| Family | Rare and salivary gland-type tumours: Introduction |
| Type | Secretory carcinoma |
| Subtype(s) | N/A |
WHO Essential and Desirable Genetic Diagnostic Criteria
| WHO Essential Criteria (Genetics)* | |
| WHO Desirable Criteria (Genetics)* | ETV6::NTRK3 fusion |
| Other Classification |
*Note: These are only the genetic/genomic criteria. Additional diagnostic criteria can be found in the WHO Classification of Tumours.
Related Terminology
| Acceptable | |
| Not Recommended |
Gene Rearrangements
| Driver Gene | Fusion(s) and Common Partner Genes | Molecular Pathogenesis | Typical Chromosomal Alteration(s) | Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|---|
| NTRK3 | ETV6::NTRK3[1] | Fusion results in constitutive activation of NTRK3 tyrosine kinase | t(12;15)(p13;q25) | Common | D, P, T | The ETV6::NTRK3 fusion is diagnostic of secretory carcinoma in the appropriate morphologic and clinical context.[2][3][4] This fusion is responsive to TRK inhibitor therapies such as larotrectinib abd entrectinib. | |
Individual Region Genomic Gain/Loss/LOH
| Chr # | Gain, Loss, Amp, LOH | Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size] | Relevant Gene(s) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
Characteristic Chromosomal or Other Global Mutational Patterns
| Chromosomal Pattern | Molecular Pathogenesis | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|
Gene Mutations (SNV/INDEL)
| Gene | Genetic Alteration | Tumor Suppressor Gene, Oncogene, Other | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
Note: A more extensive list of mutations can be found in cBioportal, COSMIC, and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
Epigenomic Alterations
Genes and Main Pathways Involved
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| NTRK3; Activating fusion with 5' partner ETV6 | MAPK/PI3K/AKT signaling | Increased cell growth and proliferation |
Genetic Diagnostic Testing Methods
FISH, RT-PCR, next generation sequencing
Familial Forms
None
Additional Information
Links
https://www.pathologyoutlines.com/topic/breastmalignantjuvenile.html
Notes
Prior Author(s):
References
- ↑ Tognon, Cristina; et al. (2002-11). "Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma". Cancer Cell. 2 (5): 367–376. doi:10.1016/s1535-6108(02)00180-0. ISSN 1535-6108. PMID 12450792. Check date values in:
|date=(help) - ↑ Arce, C.; et al. (2005-06-17). "Secretory carcinoma of the breast containing the ETV6-NTRK3 fusion gene in a male: case report and review of the literature". World Journal of Surgical Oncology. 3: 35. doi:10.1186/1477-7819-3-35. ISSN 1477-7819. PMC 1184104. PMID 15963235.
- ↑ Jacob, John Doromal; et al. (2016-06). "Rare breast cancer: 246 invasive secretory carcinomas from the National Cancer Data Base". Journal of Surgical Oncology. 113 (7): 721–725. doi:10.1002/jso.24241. ISSN 1096-9098. PMID 27040042. Check date values in:
|date=(help) - ↑ Li, Dali; et al. (2012-04). "Secretory breast carcinoma: a clinicopathological and immunophenotypic study of 15 cases with a review of the literature". Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc. 25 (4): 567–575. doi:10.1038/modpathol.2011.190. ISSN 1530-0285. PMID 22157932. Check date values in:
|date=(help)
*Citation of this Page: “Secretory carcinoma”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 04/16/2025, https://ccga.io/index.php/BRST5:Secretory carcinoma.